Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
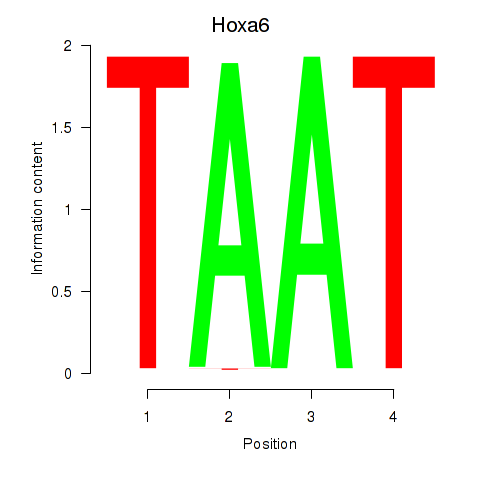
Results for Hoxa6
Z-value: 1.33
Transcription factors associated with Hoxa6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa6
|
ENSMUSG00000043219.10 | Hoxa6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa6 | mm39_v1_chr6_-_52185674_52185702 | -0.03 | 8.7e-01 | Click! |
Activity profile of Hoxa6 motif
Sorted Z-values of Hoxa6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_87695117 | 3.38 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr14_-_118289557 | 3.23 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr2_+_70305267 | 3.20 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr1_-_150341911 | 2.98 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chrM_+_5319 | 2.77 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr7_+_51537645 | 2.67 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr6_+_15185202 | 2.37 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr10_+_87694924 | 2.30 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr9_-_44714263 | 2.28 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr14_+_28740162 | 2.21 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr2_-_18053595 | 2.07 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr5_+_42225303 | 2.03 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr12_-_104439589 | 2.03 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr16_+_42727926 | 1.99 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_-_138056914 | 1.94 |
ENSMUST00000171804.4
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr6_-_138404076 | 1.94 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chrM_+_7006 | 1.77 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_-_37472290 | 1.74 |
ENSMUST00000176205.8
|
Zfp536
|
zinc finger protein 536 |
| chr4_+_11579648 | 1.74 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr12_-_25147139 | 1.70 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr1_-_165830160 | 1.66 |
ENSMUST00000111429.11
ENSMUST00000176800.2 ENSMUST00000177358.8 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chrM_+_8603 | 1.60 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr14_+_75693396 | 1.50 |
ENSMUST00000164848.3
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr3_+_29568055 | 1.49 |
ENSMUST00000140288.2
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr11_+_77821626 | 1.42 |
ENSMUST00000093995.10
ENSMUST00000000646.14 |
Sez6
|
seizure related gene 6 |
| chr8_-_85500010 | 1.39 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr2_+_153334710 | 1.38 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr13_-_53627110 | 1.38 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr7_-_37472979 | 1.35 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr11_+_96177449 | 1.35 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7 |
| chr5_+_90608751 | 1.34 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chrM_+_10167 | 1.32 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr12_-_31763859 | 1.31 |
ENSMUST00000057783.6
ENSMUST00000236002.2 ENSMUST00000174480.3 ENSMUST00000176710.2 |
Gpr22
|
G protein-coupled receptor 22 |
| chr1_-_14374794 | 1.31 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr4_+_13751297 | 1.31 |
ENSMUST00000105566.9
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr8_+_91681898 | 1.30 |
ENSMUST00000209746.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_49408040 | 1.30 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr1_-_163141278 | 1.30 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr13_+_83672389 | 1.28 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr15_-_100497863 | 1.27 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr16_+_43323970 | 1.26 |
ENSMUST00000126100.8
ENSMUST00000123047.8 ENSMUST00000156981.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_+_6800275 | 1.26 |
ENSMUST00000043578.13
ENSMUST00000139756.8 ENSMUST00000131467.8 ENSMUST00000150761.8 ENSMUST00000151281.8 |
St18
|
suppression of tumorigenicity 18 |
| chr1_-_14374842 | 1.25 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr18_-_81029986 | 1.23 |
ENSMUST00000057950.9
|
Sall3
|
spalt like transcription factor 3 |
| chr4_-_82623972 | 1.22 |
ENSMUST00000155821.2
|
Nfib
|
nuclear factor I/B |
| chr6_+_15185562 | 1.18 |
ENSMUST00000137628.8
|
Foxp2
|
forkhead box P2 |
| chr6_+_15184630 | 1.17 |
ENSMUST00000115470.3
|
Foxp2
|
forkhead box P2 |
| chr2_-_18053158 | 1.12 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1 |
| chr8_-_5155347 | 1.12 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr11_-_33153587 | 1.12 |
ENSMUST00000037746.8
|
Tlx3
|
T cell leukemia, homeobox 3 |
| chr4_-_21685781 | 1.08 |
ENSMUST00000076206.11
|
Prdm13
|
PR domain containing 13 |
| chr4_-_14621669 | 1.05 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr14_-_48900192 | 1.03 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr16_+_43184191 | 1.03 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_97666279 | 1.02 |
ENSMUST00000146447.8
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr13_+_95012107 | 1.02 |
ENSMUST00000022195.13
|
Otp
|
orthopedia homeobox |
| chr11_-_99213769 | 1.02 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr2_+_61634797 | 1.02 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr18_-_14105591 | 1.02 |
ENSMUST00000234025.2
|
Zfp521
|
zinc finger protein 521 |
| chr15_-_50753437 | 1.02 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr4_+_97665992 | 1.01 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr10_+_90412432 | 1.01 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_+_19282278 | 1.00 |
ENSMUST00000064976.6
|
Tfap2b
|
transcription factor AP-2 beta |
| chr8_+_48275178 | 0.98 |
ENSMUST00000079639.3
|
Cldn24
|
claudin 24 |
| chr10_+_85763545 | 0.97 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr6_+_15185399 | 0.96 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr4_+_99184137 | 0.96 |
ENSMUST00000094955.3
|
Gm12689
|
predicted gene 12689 |
| chr17_-_45125537 | 0.95 |
ENSMUST00000113571.10
|
Runx2
|
runt related transcription factor 2 |
| chr7_+_131144596 | 0.95 |
ENSMUST00000046093.6
|
Hmx3
|
H6 homeobox 3 |
| chr5_+_115373895 | 0.95 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr13_-_116446166 | 0.94 |
ENSMUST00000036060.13
|
Isl1
|
ISL1 transcription factor, LIM/homeodomain |
| chr15_+_92495007 | 0.92 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr2_-_7400690 | 0.92 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr14_-_109151590 | 0.91 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr11_+_96173355 | 0.91 |
ENSMUST00000125410.2
|
Hoxb8
|
homeobox B8 |
| chr18_-_81029751 | 0.91 |
ENSMUST00000238808.2
|
Sall3
|
spalt like transcription factor 3 |
| chr13_+_83672654 | 0.90 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_57010921 | 0.90 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr16_+_43067641 | 0.89 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_14621805 | 0.89 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_-_13839914 | 0.88 |
ENSMUST00000060442.14
|
Gpr85
|
G protein-coupled receptor 85 |
| chr6_+_104469751 | 0.88 |
ENSMUST00000161446.2
ENSMUST00000161070.8 ENSMUST00000089215.12 |
Cntn6
|
contactin 6 |
| chr10_+_90412114 | 0.88 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_71359000 | 0.87 |
ENSMUST00000126400.2
|
Dlx1
|
distal-less homeobox 1 |
| chr19_+_56276343 | 0.87 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr13_+_83672708 | 0.87 |
ENSMUST00000199105.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr12_-_56660054 | 0.86 |
ENSMUST00000072631.6
|
Nkx2-9
|
NK2 homeobox 9 |
| chr4_+_114678919 | 0.86 |
ENSMUST00000137570.2
|
Gm12830
|
predicted gene 12830 |
| chr2_+_74557418 | 0.85 |
ENSMUST00000111980.4
|
Hoxd4
|
homeobox D4 |
| chr8_+_121854566 | 0.85 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr14_-_124914516 | 0.85 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr5_+_90666791 | 0.84 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr2_-_35994819 | 0.82 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr10_+_90412638 | 0.81 |
ENSMUST00000183136.8
ENSMUST00000182595.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_+_69726055 | 0.81 |
ENSMUST00000114978.9
|
Tcf4
|
transcription factor 4 |
| chr5_-_67256388 | 0.81 |
ENSMUST00000174251.2
|
Phox2b
|
paired-like homeobox 2b |
| chr11_-_99412084 | 0.81 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr6_-_93890237 | 0.80 |
ENSMUST00000204167.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr7_-_49286594 | 0.80 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr6_-_52203146 | 0.80 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr15_-_50753061 | 0.79 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr17_-_85995680 | 0.79 |
ENSMUST00000024947.8
ENSMUST00000163568.4 |
Six2
|
sine oculis-related homeobox 2 |
| chr2_+_74522258 | 0.78 |
ENSMUST00000061745.5
|
Hoxd10
|
homeobox D10 |
| chr10_+_90412539 | 0.78 |
ENSMUST00000182284.8
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr17_+_85928459 | 0.78 |
ENSMUST00000162695.3
|
Six3
|
sine oculis-related homeobox 3 |
| chr4_-_110144676 | 0.77 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr11_+_42312150 | 0.77 |
ENSMUST00000192403.2
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr10_+_90412570 | 0.77 |
ENSMUST00000182430.8
ENSMUST00000182960.8 ENSMUST00000182045.2 ENSMUST00000182083.2 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr19_+_22670134 | 0.76 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr19_-_45224251 | 0.75 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr18_-_79152504 | 0.75 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr5_-_66238313 | 0.75 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr11_-_99412162 | 0.74 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr1_-_13061333 | 0.74 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr7_-_37469049 | 0.74 |
ENSMUST00000175941.8
|
Zfp536
|
zinc finger protein 536 |
| chr1_-_163141230 | 0.73 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chr13_-_40883893 | 0.73 |
ENSMUST00000021787.7
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr2_+_79538124 | 0.72 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr2_+_181408833 | 0.71 |
ENSMUST00000108756.8
|
Myt1
|
myelin transcription factor 1 |
| chr13_+_83652150 | 0.70 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr5_-_142803135 | 0.70 |
ENSMUST00000198181.2
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr9_-_71070506 | 0.69 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr5_-_67256578 | 0.69 |
ENSMUST00000012664.11
|
Phox2b
|
paired-like homeobox 2b |
| chr4_-_97472844 | 0.69 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr5_+_139529643 | 0.69 |
ENSMUST00000174792.2
|
Uncx
|
UNC homeobox |
| chr6_+_104470181 | 0.68 |
ENSMUST00000162872.2
|
Cntn6
|
contactin 6 |
| chr13_+_83652280 | 0.68 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_96183294 | 0.68 |
ENSMUST00000173432.3
|
Hoxb6
|
homeobox B6 |
| chr6_-_136148820 | 0.68 |
ENSMUST00000188999.3
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr2_-_63014514 | 0.68 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr17_-_45125468 | 0.68 |
ENSMUST00000159943.8
ENSMUST00000160673.8 |
Runx2
|
runt related transcription factor 2 |
| chr2_+_74528071 | 0.67 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9 |
| chr2_+_9887427 | 0.67 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr1_-_164988342 | 0.67 |
ENSMUST00000027859.12
|
Tbx19
|
T-box 19 |
| chr2_+_105505772 | 0.66 |
ENSMUST00000111085.8
|
Pax6
|
paired box 6 |
| chr13_+_42834989 | 0.66 |
ENSMUST00000149235.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr9_+_44309727 | 0.66 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr14_-_30213408 | 0.65 |
ENSMUST00000112250.6
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr1_+_66361252 | 0.65 |
ENSMUST00000123647.8
|
Map2
|
microtubule-associated protein 2 |
| chr2_-_63014622 | 0.64 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_+_109522781 | 0.64 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr1_+_81055201 | 0.63 |
ENSMUST00000123285.2
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr11_+_96173475 | 0.63 |
ENSMUST00000168043.2
|
Hoxb8
|
homeobox B8 |
| chr5_-_147244074 | 0.63 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr14_+_32321341 | 0.62 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr10_+_42736539 | 0.62 |
ENSMUST00000157071.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr16_+_43184507 | 0.61 |
ENSMUST00000148775.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr5_+_20433169 | 0.61 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr18_-_23174698 | 0.60 |
ENSMUST00000097651.10
|
Nol4
|
nucleolar protein 4 |
| chr2_+_83786742 | 0.60 |
ENSMUST00000178325.2
|
Gm13698
|
predicted gene 13698 |
| chrM_+_2743 | 0.60 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr2_+_65760477 | 0.60 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr10_-_42459624 | 0.60 |
ENSMUST00000019938.11
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr6_+_79995860 | 0.59 |
ENSMUST00000147663.8
ENSMUST00000128718.8 ENSMUST00000126005.8 ENSMUST00000133918.8 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr19_+_56276375 | 0.59 |
ENSMUST00000166049.8
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr6_+_53264255 | 0.59 |
ENSMUST00000203528.3
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr17_+_44263890 | 0.58 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr4_-_14621497 | 0.58 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_+_79995994 | 0.58 |
ENSMUST00000126399.2
ENSMUST00000136421.2 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr5_+_115573055 | 0.58 |
ENSMUST00000136586.6
|
Msi1
|
musashi RNA-binding protein 1 |
| chr4_-_110149916 | 0.57 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr14_+_33662976 | 0.57 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr15_+_18819033 | 0.57 |
ENSMUST00000166873.9
|
Cdh10
|
cadherin 10 |
| chr5_+_67125902 | 0.56 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr3_-_37180093 | 0.56 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr5_+_118307754 | 0.56 |
ENSMUST00000054836.7
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr16_+_43056218 | 0.56 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_-_87613301 | 0.55 |
ENSMUST00000034991.8
|
Tbx18
|
T-box18 |
| chr1_-_87322443 | 0.55 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr18_+_82929037 | 0.55 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr6_-_13838423 | 0.55 |
ENSMUST00000115492.2
|
Gpr85
|
G protein-coupled receptor 85 |
| chr3_+_18108313 | 0.55 |
ENSMUST00000026120.8
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr2_+_181409075 | 0.55 |
ENSMUST00000108757.9
|
Myt1
|
myelin transcription factor 1 |
| chr5_-_24732200 | 0.55 |
ENSMUST00000088311.6
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr1_+_134217727 | 0.54 |
ENSMUST00000027730.6
|
Myog
|
myogenin |
| chr6_-_138398376 | 0.54 |
ENSMUST00000163065.8
|
Lmo3
|
LIM domain only 3 |
| chr9_+_118307412 | 0.53 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr2_+_105505823 | 0.53 |
ENSMUST00000167211.9
ENSMUST00000111083.10 |
Pax6
|
paired box 6 |
| chr5_+_88635834 | 0.53 |
ENSMUST00000199104.5
ENSMUST00000031222.9 |
Enam
|
enamelin |
| chr5_+_104447037 | 0.53 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr9_-_51076724 | 0.53 |
ENSMUST00000210433.2
|
Gm32742
|
predicted gene, 32742 |
| chr17_+_93506435 | 0.53 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr15_+_102927366 | 0.53 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr1_+_6805048 | 0.52 |
ENSMUST00000139838.8
|
St18
|
suppression of tumorigenicity 18 |
| chr3_-_137837117 | 0.52 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr9_-_112016966 | 0.52 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr9_-_48876290 | 0.51 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr4_-_58499398 | 0.51 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr12_+_29578354 | 0.51 |
ENSMUST00000218583.2
ENSMUST00000049784.17 |
Myt1l
|
myelin transcription factor 1-like |
| chr11_-_18968714 | 0.51 |
ENSMUST00000177417.8
|
Meis1
|
Meis homeobox 1 |
| chr11_+_96162283 | 0.50 |
ENSMUST00000000010.9
ENSMUST00000174042.3 |
Hoxb9
|
homeobox B9 |
| chr2_-_32976378 | 0.50 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr7_+_49559859 | 0.50 |
ENSMUST00000056442.12
|
Slc6a5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
| chr13_-_40887244 | 0.49 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr16_-_26810402 | 0.49 |
ENSMUST00000231299.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr9_-_37580478 | 0.49 |
ENSMUST00000011262.4
|
Panx3
|
pannexin 3 |
| chr2_+_97298002 | 0.49 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr3_+_55369384 | 0.49 |
ENSMUST00000200352.2
|
Dclk1
|
doublecortin-like kinase 1 |
| chr10_+_52267702 | 0.48 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr6_-_119940694 | 0.48 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.8 | 5.7 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.6 | 2.4 | GO:0048880 | sensory system development(GO:0048880) |
| 0.6 | 1.7 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.5 | 1.4 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.4 | 1.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 4.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.4 | 1.1 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.3 | 1.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 1.0 | GO:0097276 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.3 | 3.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.3 | 4.9 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 | 4.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 1.6 | GO:0003409 | optic vesicle morphogenesis(GO:0003404) optic cup structural organization(GO:0003409) |
| 0.3 | 3.7 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 1.2 | GO:0061141 | lung ciliated cell differentiation(GO:0061141) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 1.5 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.3 | 1.2 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.3 | 0.9 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.3 | 2.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.6 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.3 | 2.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 1.2 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.2 | 1.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 0.8 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 0.6 | GO:0061193 | taste bud development(GO:0061193) |
| 0.2 | 0.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 0.4 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.2 | 1.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 0.9 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.2 | 1.5 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 2.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 0.5 | GO:0019230 | proprioception(GO:0019230) sensory neuron axon guidance(GO:0097374) |
| 0.2 | 0.5 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.2 | 2.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 0.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 2.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.7 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 1.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 3.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 3.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.7 | GO:0021889 | olfactory bulb interneuron differentiation(GO:0021889) |
| 0.1 | 1.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.7 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.4 | GO:1904501 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) dorsal root ganglion development(GO:1990791) |
| 0.1 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.5 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.5 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 6.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.0 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.1 | 1.0 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.8 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 3.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.4 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 1.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 1.4 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.7 | GO:0097090 | presynaptic membrane organization(GO:0097090) |
| 0.1 | 1.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.3 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.6 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 3.4 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 1.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.5 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 2.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.4 | GO:0071638 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.2 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.2 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.4 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.4 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 3.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.8 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.4 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 1.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.6 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 1.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.7 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 2.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.7 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.6 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.6 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.4 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 1.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.9 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.0 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.2 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 1.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 1.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 4.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.7 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.2 | 0.5 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.2 | 0.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.6 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 1.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 3.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 1.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.6 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 5.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 2.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 0.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 4.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.7 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.2 | 0.7 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 6.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 6.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 3.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 6.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 2.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.7 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.1 | 5.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 3.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.0 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) |
| 0.1 | 0.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.8 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 4.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 2.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.9 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 23.3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.5 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.1 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 1.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.4 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 4.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 4.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.9 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 1.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.9 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 2.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 2.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |