Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
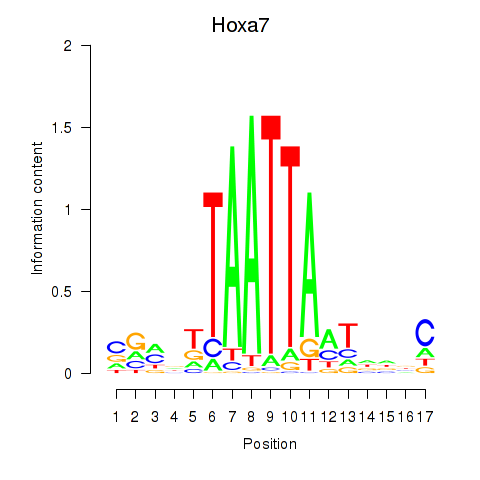
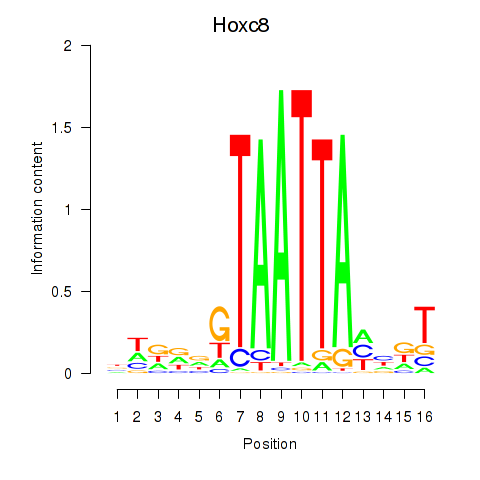
Results for Hoxa7_Hoxc8
Z-value: 0.56
Transcription factors associated with Hoxa7_Hoxc8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa7
|
ENSMUSG00000038236.9 | Hoxa7 |
|
Hoxc8
|
ENSMUSG00000001657.8 | Hoxc8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa7 | mm39_v1_chr6_-_52194440_52194485 | 0.29 | 8.2e-02 | Click! |
| Hoxc8 | mm39_v1_chr15_+_102898966_102899039 | 0.10 | 5.8e-01 | Click! |
Activity profile of Hoxa7_Hoxc8 motif
Sorted Z-values of Hoxa7_Hoxc8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa7_Hoxc8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_30541581 | 3.00 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr14_+_80237691 | 2.99 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr2_+_36120438 | 1.21 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr2_+_174292471 | 1.12 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr5_-_116162415 | 0.83 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr6_-_137146708 | 0.61 |
ENSMUST00000117919.8
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr7_+_30193047 | 0.61 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr1_-_134883645 | 0.58 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr8_+_46338498 | 0.58 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr6_-_115014777 | 0.55 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr10_+_97318223 | 0.54 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr4_+_34893772 | 0.54 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr18_+_23886765 | 0.52 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_-_37014859 | 0.44 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr3_-_75177378 | 0.43 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr10_-_45346297 | 0.43 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr8_+_23901506 | 0.42 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr2_-_113678999 | 0.42 |
ENSMUST00000102545.8
ENSMUST00000110948.8 |
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr14_+_54669054 | 0.41 |
ENSMUST00000089688.6
ENSMUST00000225641.2 |
Mmp14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr3_+_57332735 | 0.40 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr8_+_46338557 | 0.39 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr2_-_84255602 | 0.38 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr12_-_40298072 | 0.38 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr5_-_65855511 | 0.38 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr19_+_5524701 | 0.37 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr9_+_21437440 | 0.36 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chrX_+_164953444 | 0.35 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr9_+_113641615 | 0.35 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr19_+_34078333 | 0.34 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr16_-_75706161 | 0.32 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr19_-_53020531 | 0.31 |
ENSMUST00000236008.2
ENSMUST00000237294.2 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr10_-_128462616 | 0.31 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr6_+_37847721 | 0.30 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr4_-_136329953 | 0.29 |
ENSMUST00000105847.8
ENSMUST00000116273.9 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr18_-_43610829 | 0.29 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr4_-_43710231 | 0.29 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr12_-_55061117 | 0.29 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr3_-_66204228 | 0.28 |
ENSMUST00000029419.8
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr9_-_58648826 | 0.28 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr13_-_55169000 | 0.28 |
ENSMUST00000153665.8
|
Hk3
|
hexokinase 3 |
| chr1_-_171854818 | 0.27 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr14_+_26722319 | 0.26 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr9_-_119897328 | 0.24 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr9_-_119897358 | 0.23 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr5_-_87716882 | 0.23 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr19_-_53932581 | 0.22 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr1_+_82817794 | 0.22 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr1_-_4479445 | 0.21 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr7_-_103094646 | 0.21 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr8_-_49008305 | 0.20 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr19_-_39875192 | 0.19 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr6_-_122317484 | 0.19 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr15_-_82678490 | 0.19 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr2_-_45007407 | 0.19 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_102446883 | 0.19 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr19_-_32173824 | 0.17 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr10_+_102376109 | 0.16 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr2_+_83554741 | 0.15 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr2_+_69050315 | 0.15 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr11_-_99265721 | 0.14 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chrX_+_158086253 | 0.14 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr3_-_59127571 | 0.14 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr16_-_19132814 | 0.13 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chr7_+_45271229 | 0.13 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr6_+_48723122 | 0.12 |
ENSMUST00000055558.6
|
Gimap5
|
GTPase, IMAP family member 5 |
| chr18_+_31742565 | 0.12 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr18_+_4920513 | 0.12 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr11_-_73382303 | 0.11 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr14_+_65612788 | 0.11 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chrX_+_9751861 | 0.11 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr7_-_103778992 | 0.11 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr3_-_72875187 | 0.11 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr7_-_37718916 | 0.10 |
ENSMUST00000085513.6
ENSMUST00000206327.2 |
Uri1
|
URI1, prefoldin-like chaperone |
| chr2_+_85868891 | 0.10 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr19_-_53932867 | 0.10 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr5_-_62923463 | 0.10 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr16_-_56688024 | 0.10 |
ENSMUST00000232373.2
|
Tmem45a
|
transmembrane protein 45a |
| chr16_+_45044678 | 0.10 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr18_+_88989914 | 0.09 |
ENSMUST00000023828.9
|
Rttn
|
rotatin |
| chr19_-_38807600 | 0.09 |
ENSMUST00000025963.8
|
Noc3l
|
NOC3 like DNA replication regulator |
| chr17_-_29226700 | 0.09 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38 |
| chr10_-_75946790 | 0.09 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr11_-_79418500 | 0.08 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr19_+_13339600 | 0.08 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467 |
| chr7_+_84502761 | 0.08 |
ENSMUST00000217039.3
ENSMUST00000211582.2 |
Olfr291
|
olfactory receptor 291 |
| chr2_-_86109346 | 0.08 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr2_+_111581351 | 0.07 |
ENSMUST00000207590.4
|
Olfr1301
|
olfactory receptor 1301 |
| chr9_+_109881083 | 0.07 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chr4_+_145311759 | 0.07 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr18_+_32200781 | 0.06 |
ENSMUST00000025243.5
ENSMUST00000212675.2 |
Iws1
|
IWS1, SUPT6 interacting protein |
| chr5_-_86616849 | 0.06 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr19_+_55883924 | 0.06 |
ENSMUST00000111646.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr2_+_83554868 | 0.06 |
ENSMUST00000111740.9
|
Itgav
|
integrin alpha V |
| chr13_-_32967937 | 0.06 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr19_-_41921676 | 0.06 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr16_+_35803794 | 0.06 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr15_-_100321973 | 0.05 |
ENSMUST00000154676.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr3_+_32490300 | 0.05 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr2_-_85632888 | 0.05 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr9_-_79884920 | 0.05 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr9_-_79885063 | 0.05 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr14_-_36641270 | 0.04 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr16_-_19241884 | 0.04 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr9_+_39985125 | 0.04 |
ENSMUST00000054051.5
|
Olfr982
|
olfactory receptor 982 |
| chr3_+_5283606 | 0.04 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr18_+_37651393 | 0.03 |
ENSMUST00000097609.3
|
Pcdhb22
|
protocadherin beta 22 |
| chr17_-_29226886 | 0.03 |
ENSMUST00000232723.2
|
Stk38
|
serine/threonine kinase 38 |
| chr16_+_13176238 | 0.03 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr14_-_14255736 | 0.03 |
ENSMUST00000170111.3
|
Kctd6
|
potassium channel tetramerisation domain containing 6 |
| chr10_-_44024843 | 0.02 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr3_+_5283577 | 0.02 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr1_-_4563821 | 0.02 |
ENSMUST00000191939.2
|
Sox17
|
SRY (sex determining region Y)-box 17 |
| chr2_+_85648823 | 0.02 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr2_-_87504008 | 0.02 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr1_-_134883577 | 0.02 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr2_+_88470886 | 0.02 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr10_-_20600442 | 0.02 |
ENSMUST00000170265.8
|
Pde7b
|
phosphodiesterase 7B |
| chr19_-_53933052 | 0.02 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr17_-_29226965 | 0.02 |
ENSMUST00000009138.13
ENSMUST00000119274.3 |
Stk38
|
serine/threonine kinase 38 |
| chr11_-_73348284 | 0.02 |
ENSMUST00000121209.3
ENSMUST00000127789.3 |
Olfr380
|
olfactory receptor 380 |
| chr7_-_107696793 | 0.01 |
ENSMUST00000217304.2
|
Olfr482
|
olfactory receptor 482 |
| chr2_+_110427643 | 0.01 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr15_+_25774070 | 0.01 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr10_+_115405891 | 0.01 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr18_+_37827413 | 0.01 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr2_-_86016027 | 0.01 |
ENSMUST00000215138.3
|
Olfr52
|
olfactory receptor 52 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.2 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 0.5 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 0.4 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 0.4 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 3.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.4 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.4 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.3 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.5 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 3.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.3 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |