Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
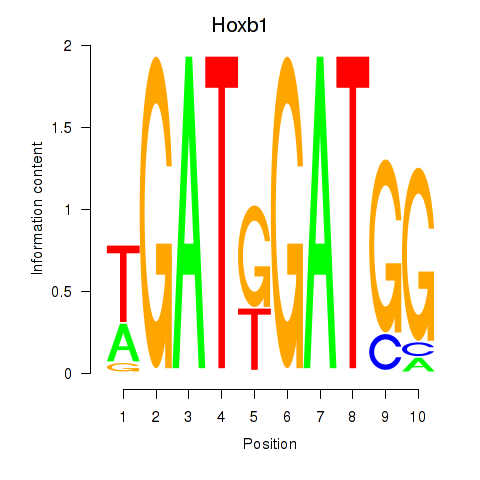
Results for Hoxb1
Z-value: 1.06
Transcription factors associated with Hoxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb1
|
ENSMUSG00000018973.3 | Hoxb1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb1 | mm39_v1_chr11_+_96256565_96256578 | -0.28 | 1.0e-01 | Click! |
Activity profile of Hoxb1 motif
Sorted Z-values of Hoxb1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_97066937 | 5.77 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr7_+_123061535 | 2.27 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr7_+_123061497 | 2.25 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr4_+_118285275 | 1.64 |
ENSMUST00000006557.13
ENSMUST00000167636.8 ENSMUST00000102673.11 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr18_-_56695333 | 1.30 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr6_-_124441731 | 1.19 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr18_-_56695259 | 1.07 |
ENSMUST00000171844.3
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr18_-_56695288 | 0.98 |
ENSMUST00000170309.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr15_+_74388044 | 0.93 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr11_-_97913420 | 0.92 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr5_+_42225303 | 0.74 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr7_+_100970910 | 0.70 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr14_+_70314652 | 0.69 |
ENSMUST00000035908.3
|
Egr3
|
early growth response 3 |
| chr10_-_33500583 | 0.69 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr7_+_100971034 | 0.66 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr13_-_21685588 | 0.65 |
ENSMUST00000044043.3
|
Cox5b-ps
|
cytochrome c oxidase subunit 5B, pseudogene |
| chr4_-_102971434 | 0.64 |
ENSMUST00000036557.15
ENSMUST00000036451.15 |
Dnai4
|
dynein axonemal intermediate chain 4 |
| chr4_-_102971752 | 0.63 |
ENSMUST00000106868.4
|
Dnai4
|
dynein axonemal intermediate chain 4 |
| chrX_+_109857866 | 0.62 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr10_-_67120959 | 0.60 |
ENSMUST00000159002.2
ENSMUST00000077839.13 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chr11_+_76563281 | 0.58 |
ENSMUST00000056184.2
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr11_+_96256565 | 0.56 |
ENSMUST00000019117.3
|
Hoxb1
|
homeobox B1 |
| chr4_+_102971909 | 0.55 |
ENSMUST00000143417.8
|
Mier1
|
MEIR1 treanscription regulator |
| chr11_+_98277276 | 0.48 |
ENSMUST00000041301.8
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr17_+_34866160 | 0.47 |
ENSMUST00000173984.2
|
Atf6b
|
activating transcription factor 6 beta |
| chr17_+_34866090 | 0.45 |
ENSMUST00000015605.15
|
Atf6b
|
activating transcription factor 6 beta |
| chr4_-_91288221 | 0.45 |
ENSMUST00000102799.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr9_-_91247831 | 0.45 |
ENSMUST00000065360.5
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr9_-_91247809 | 0.45 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr9_+_78303059 | 0.44 |
ENSMUST00000113367.2
|
Ddx43
|
DEAD box helicase 43 |
| chr1_+_153541020 | 0.44 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr11_-_40646090 | 0.43 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr3_-_58433313 | 0.42 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr6_-_93889483 | 0.41 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr3_+_127426783 | 0.41 |
ENSMUST00000029587.9
|
Neurog2
|
neurogenin 2 |
| chr7_+_73040908 | 0.40 |
ENSMUST00000128471.2
ENSMUST00000139780.3 |
Rgma
|
repulsive guidance molecule family member A |
| chrX_+_104807868 | 0.40 |
ENSMUST00000033581.4
|
Fgf16
|
fibroblast growth factor 16 |
| chr1_-_9770434 | 0.40 |
ENSMUST00000088658.11
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr2_+_65676111 | 0.40 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr10_-_88440869 | 0.39 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr1_-_25868788 | 0.38 |
ENSMUST00000151309.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr14_-_24054352 | 0.37 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr5_+_20112500 | 0.36 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_+_65676176 | 0.35 |
ENSMUST00000053910.10
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr10_+_36383008 | 0.33 |
ENSMUST00000168572.8
|
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chrX_+_142301666 | 0.33 |
ENSMUST00000134402.8
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr8_-_117809188 | 0.33 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr1_-_25868592 | 0.32 |
ENSMUST00000135518.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr14_-_24054273 | 0.31 |
ENSMUST00000188285.7
ENSMUST00000190044.7 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr9_+_108660989 | 0.30 |
ENSMUST00000192307.6
ENSMUST00000193560.6 ENSMUST00000194875.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr16_+_25620652 | 0.30 |
ENSMUST00000115304.8
ENSMUST00000115305.2 ENSMUST00000040231.13 ENSMUST00000115306.8 |
Trp63
|
transformation related protein 63 |
| chr18_+_66006119 | 0.29 |
ENSMUST00000025395.10
|
Grp
|
gastrin releasing peptide |
| chr14_-_24054186 | 0.29 |
ENSMUST00000188991.7
ENSMUST00000224468.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr15_-_95426419 | 0.29 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr17_+_27160203 | 0.29 |
ENSMUST00000194598.6
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr16_-_97763780 | 0.29 |
ENSMUST00000232187.2
ENSMUST00000231263.2 ENSMUST00000052089.9 ENSMUST00000063605.15 ENSMUST00000113734.9 ENSMUST00000231560.2 ENSMUST00000232165.2 |
Zbtb21
C2cd2
|
zinc finger and BTB domain containing 21 C2 calcium-dependent domain containing 2 |
| chr19_-_43376794 | 0.27 |
ENSMUST00000099428.5
|
Hpse2
|
heparanase 2 |
| chr14_-_24053994 | 0.27 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_+_142359099 | 0.27 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr11_-_69439934 | 0.26 |
ENSMUST00000108659.2
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr3_-_37286714 | 0.25 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr14_+_70314727 | 0.25 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr17_+_27160356 | 0.24 |
ENSMUST00000229490.2
ENSMUST00000201702.5 ENSMUST00000177932.7 ENSMUST00000201349.6 |
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr6_-_58884038 | 0.24 |
ENSMUST00000059539.5
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr4_+_58285957 | 0.23 |
ENSMUST00000081919.12
ENSMUST00000177951.8 ENSMUST00000098059.10 ENSMUST00000179951.2 ENSMUST00000102893.10 ENSMUST00000084578.12 ENSMUST00000098057.10 |
Musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr15_+_82230155 | 0.23 |
ENSMUST00000023086.15
|
Smdt1
|
single-pass membrane protein with aspartate rich tail 1 |
| chr5_-_124492734 | 0.22 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr8_-_88686188 | 0.22 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr19_-_41732104 | 0.22 |
ENSMUST00000025993.10
|
Slit1
|
slit guidance ligand 1 |
| chr5_+_20112704 | 0.21 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_-_28126124 | 0.21 |
ENSMUST00000115324.9
ENSMUST00000090512.10 |
Grm8
|
glutamate receptor, metabotropic 8 |
| chr10_-_88440996 | 0.21 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr15_+_98953776 | 0.20 |
ENSMUST00000229268.2
|
Prph
|
peripherin |
| chr7_+_119289249 | 0.19 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr4_+_43669266 | 0.18 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr11_+_85243970 | 0.18 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chr3_-_88669551 | 0.18 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr8_+_108669276 | 0.17 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr4_+_109092829 | 0.16 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chr9_-_107960528 | 0.16 |
ENSMUST00000159372.3
ENSMUST00000160249.8 |
Rnf123
|
ring finger protein 123 |
| chr8_-_118400418 | 0.13 |
ENSMUST00000173522.8
ENSMUST00000174450.2 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr19_-_5553804 | 0.12 |
ENSMUST00000189704.2
|
Nscme3l
|
NSE3 homolog, SMC5-SMC6 complex component like |
| chr11_-_102187445 | 0.12 |
ENSMUST00000107132.3
ENSMUST00000073234.9 |
Atxn7l3
|
ataxin 7-like 3 |
| chr15_+_38740784 | 0.12 |
ENSMUST00000226440.3
ENSMUST00000239553.1 |
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr17_+_87224776 | 0.12 |
ENSMUST00000042172.7
|
Tmem247
|
transmembrane protein 247 |
| chr12_+_38830081 | 0.12 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr10_-_12745109 | 0.11 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr9_+_114917902 | 0.11 |
ENSMUST00000182363.8
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr14_+_50656083 | 0.11 |
ENSMUST00000216949.2
|
Olfr739
|
olfactory receptor 739 |
| chr17_-_23939700 | 0.10 |
ENSMUST00000201734.4
|
1520401A03Rik
|
RIKEN cDNA 1520401A03 gene |
| chr4_+_109092610 | 0.10 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr1_-_135934080 | 0.09 |
ENSMUST00000166193.9
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr1_+_178626003 | 0.09 |
ENSMUST00000160789.2
|
Kif26b
|
kinesin family member 26B |
| chr2_+_156317416 | 0.08 |
ENSMUST00000029155.16
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr2_-_90900525 | 0.06 |
ENSMUST00000153367.2
ENSMUST00000079976.10 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr3_-_7678785 | 0.05 |
ENSMUST00000194279.6
|
Il7
|
interleukin 7 |
| chr10_+_81464536 | 0.05 |
ENSMUST00000129622.2
|
Ankrd24
|
ankyrin repeat domain 24 |
| chr2_-_90900628 | 0.04 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr3_-_7678796 | 0.03 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr19_-_47452557 | 0.02 |
ENSMUST00000111800.4
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chrX_+_165127688 | 0.02 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr5_-_138270995 | 0.00 |
ENSMUST00000161665.2
ENSMUST00000100530.8 ENSMUST00000161279.8 ENSMUST00000161647.8 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 1.2 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.2 | 1.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.7 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 5.8 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.1 | 1.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0060197 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 0.1 | 0.5 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 0.6 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.1 | 1.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.4 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 0.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.9 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.3 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.3 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.5 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.6 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.4 | GO:1900121 | negative regulation of axon regeneration(GO:0048681) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.4 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 1.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.6 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.3 | 4.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 3.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |