Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxb8_Pdx1
Z-value: 2.13
Transcription factors associated with Hoxb8_Pdx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
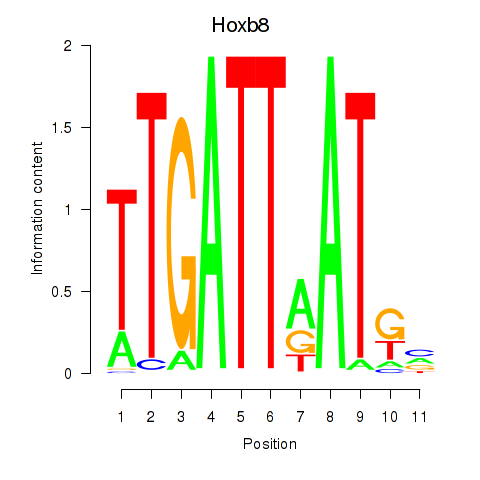
Hoxb8
|
ENSMUSG00000056648.6 | Hoxb8 |
|
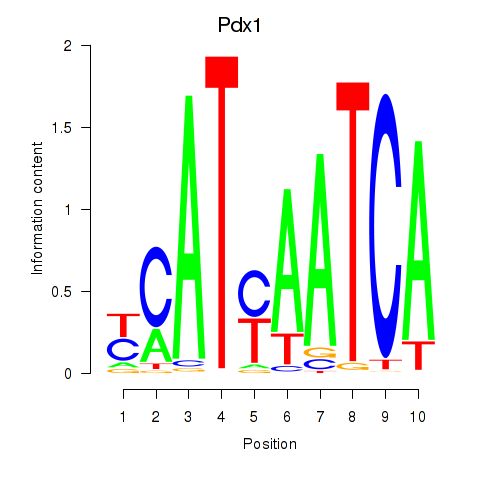
Pdx1
|
ENSMUSG00000029644.8 | Pdx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb8 | mm39_v1_chr11_+_96173475_96173551 | -0.25 | 1.5e-01 | Click! |
| Pdx1 | mm39_v1_chr5_+_147206769_147206912 | -0.21 | 2.2e-01 | Click! |
Activity profile of Hoxb8_Pdx1 motif
Sorted Z-values of Hoxb8_Pdx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb8_Pdx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_130754413 | 19.72 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr13_-_24098981 | 11.63 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr7_+_130633776 | 11.59 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr13_-_24098951 | 11.00 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr15_+_31224616 | 7.64 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr15_+_31225302 | 7.34 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr5_-_87074380 | 7.12 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr3_-_20329823 | 7.06 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr13_+_4109566 | 6.80 |
ENSMUST00000041768.7
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr19_+_30210320 | 6.37 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr15_+_31224555 | 5.85 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr7_-_46392403 | 5.83 |
ENSMUST00000128088.4
|
Saa1
|
serum amyloid A 1 |
| chr19_+_4761181 | 5.65 |
ENSMUST00000008991.8
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr5_-_87288177 | 5.37 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr19_+_39980868 | 5.28 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr15_+_31224460 | 5.08 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr14_+_55798362 | 4.79 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr1_+_88062508 | 4.71 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr14_+_55797934 | 4.66 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr7_+_46401214 | 4.55 |
ENSMUST00000210769.2
ENSMUST00000210272.2 ENSMUST00000075982.4 |
Saa2
|
serum amyloid A 2 |
| chr5_-_87572060 | 4.53 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr6_+_128639342 | 4.51 |
ENSMUST00000032518.7
ENSMUST00000204416.2 |
Clec2h
|
C-type lectin domain family 2, member h |
| chr1_+_88030951 | 4.33 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr8_-_95422851 | 4.20 |
ENSMUST00000034227.6
|
Pllp
|
plasma membrane proteolipid |
| chr2_-_84605764 | 3.81 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr16_+_22769822 | 3.76 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_22769844 | 3.53 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr2_-_84605732 | 3.51 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr5_-_130053120 | 3.44 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr1_-_162694076 | 3.43 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr14_+_55797443 | 3.42 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr11_-_84058292 | 3.40 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr12_-_103871146 | 3.34 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr1_-_10038030 | 3.33 |
ENSMUST00000185184.2
|
Tcf24
|
transcription factor 24 |
| chr13_+_23991010 | 3.27 |
ENSMUST00000006786.11
ENSMUST00000099697.3 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr14_+_55797468 | 3.25 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_-_109059414 | 3.06 |
ENSMUST00000160774.8
ENSMUST00000194478.6 ENSMUST00000030288.14 ENSMUST00000162787.9 |
Osbpl9
|
oxysterol binding protein-like 9 |
| chr12_-_103829810 | 3.03 |
ENSMUST00000085056.8
ENSMUST00000072876.12 ENSMUST00000124717.2 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr3_-_107850707 | 3.00 |
ENSMUST00000106681.3
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr5_-_89605622 | 2.96 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr8_-_110765983 | 2.96 |
ENSMUST00000109222.4
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr9_-_70841881 | 2.93 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr1_+_88093726 | 2.89 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr16_+_96162854 | 2.89 |
ENSMUST00000113794.8
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr11_+_102993865 | 2.79 |
ENSMUST00000152971.2
|
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr6_+_124281607 | 2.78 |
ENSMUST00000032234.5
ENSMUST00000112541.8 |
Cd163
|
CD163 antigen |
| chr18_-_35631914 | 2.73 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr10_+_21253190 | 2.71 |
ENSMUST00000042699.14
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr6_-_124519240 | 2.68 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr2_-_177567397 | 2.66 |
ENSMUST00000108934.9
ENSMUST00000081529.11 |
Zfp972
|
zinc finger protein 972 |
| chr18_-_10706701 | 2.59 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr6_-_119365632 | 2.56 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr17_-_35081129 | 2.56 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr3_-_107851021 | 2.54 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr17_-_35081456 | 2.48 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chrM_+_11735 | 2.43 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr17_-_35265514 | 2.42 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chr13_+_4099001 | 2.42 |
ENSMUST00000118717.10
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr6_-_136852792 | 2.41 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr3_-_65300000 | 2.38 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr18_-_32271224 | 2.35 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chr6_-_21851827 | 2.35 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr11_-_31621727 | 2.33 |
ENSMUST00000109415.2
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr11_-_58504307 | 2.31 |
ENSMUST00000048801.8
|
Lypd8l
|
LY6/PLAUR domain containing 8 like |
| chr11_-_49004584 | 2.30 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr3_-_157630690 | 2.26 |
ENSMUST00000118539.2
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr11_-_106471420 | 2.26 |
ENSMUST00000153870.2
|
Tex2
|
testis expressed gene 2 |
| chr16_+_90535212 | 2.21 |
ENSMUST00000038197.3
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr1_+_87983099 | 2.15 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_-_71070506 | 2.07 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr2_+_22959452 | 2.06 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr5_+_88712840 | 2.06 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr1_-_164763091 | 2.03 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr1_-_39616445 | 1.94 |
ENSMUST00000062525.11
|
Rnf149
|
ring finger protein 149 |
| chr4_-_35157405 | 1.88 |
ENSMUST00000102975.10
|
Mob3b
|
MOB kinase activator 3B |
| chr12_-_79211820 | 1.85 |
ENSMUST00000162569.8
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr10_-_41585293 | 1.85 |
ENSMUST00000019955.16
ENSMUST00000099932.10 |
Ccdc162
|
coiled-coil domain containing 162 |
| chr1_+_87983189 | 1.85 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_-_69586626 | 1.84 |
ENSMUST00000108649.3
ENSMUST00000174159.8 ENSMUST00000181810.8 |
Tnfsfm13
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, membrane-bound member 13 tumor necrosis factor (ligand) superfamily, member 12 |
| chr2_+_22958179 | 1.80 |
ENSMUST00000227663.2
ENSMUST00000028121.15 ENSMUST00000227809.2 ENSMUST00000144088.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chrX_-_36137764 | 1.76 |
ENSMUST00000047486.6
|
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr9_-_70048766 | 1.71 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr19_-_4548602 | 1.68 |
ENSMUST00000048482.8
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr6_-_84565613 | 1.62 |
ENSMUST00000204146.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr12_+_112555186 | 1.60 |
ENSMUST00000101029.4
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr6_+_37847721 | 1.59 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr14_+_74973081 | 1.59 |
ENSMUST00000177283.8
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr3_+_62327089 | 1.58 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr15_+_38869667 | 1.55 |
ENSMUST00000022906.8
|
Fzd6
|
frizzled class receptor 6 |
| chr3_-_116762617 | 1.47 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr4_+_101574601 | 1.45 |
ENSMUST00000102777.10
ENSMUST00000106921.9 ENSMUST00000037552.10 ENSMUST00000145024.2 |
Lepr
|
leptin receptor |
| chr9_-_70842090 | 1.45 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr17_+_79919267 | 1.44 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr18_+_38430015 | 1.44 |
ENSMUST00000236319.2
|
Rnf14
|
ring finger protein 14 |
| chr5_-_113369096 | 1.41 |
ENSMUST00000211733.2
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chr4_-_119217079 | 1.39 |
ENSMUST00000143494.3
ENSMUST00000154606.9 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr4_+_102446883 | 1.39 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr13_+_4283729 | 1.37 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr6_+_54244116 | 1.35 |
ENSMUST00000114402.9
|
Chn2
|
chimerin 2 |
| chr3_+_151143524 | 1.27 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr11_-_69586884 | 1.25 |
ENSMUST00000180587.8
|
Tnfsfm13
|
tumor necrosis factor (ligand) superfamily, membrane-bound member 13 |
| chr8_-_110766009 | 1.23 |
ENSMUST00000212934.2
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr7_-_12829100 | 1.22 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr10_+_121200984 | 1.21 |
ENSMUST00000040344.7
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr11_-_69586347 | 1.20 |
ENSMUST00000181261.2
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr8_+_119010458 | 1.18 |
ENSMUST00000117160.2
|
Cdh13
|
cadherin 13 |
| chr15_+_38869415 | 1.17 |
ENSMUST00000179165.9
|
Fzd6
|
frizzled class receptor 6 |
| chr4_-_150087587 | 1.14 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr18_+_38430205 | 1.14 |
ENSMUST00000235491.2
|
Rnf14
|
ring finger protein 14 |
| chr19_+_24853039 | 1.12 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr3_+_85948030 | 1.12 |
ENSMUST00000238545.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr14_-_31503869 | 1.11 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr17_+_37504783 | 1.11 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr10_+_4561974 | 1.10 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr4_+_102112189 | 1.09 |
ENSMUST00000106908.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr19_-_3625698 | 1.08 |
ENSMUST00000172362.3
ENSMUST00000025846.16 ENSMUST00000226109.2 ENSMUST00000113997.9 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr2_+_125876883 | 1.08 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr1_-_162726053 | 1.06 |
ENSMUST00000143123.3
|
Fmo2
|
flavin containing monooxygenase 2 |
| chrX_+_37689503 | 1.04 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr19_+_45064539 | 1.04 |
ENSMUST00000026234.5
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chrX_-_105055486 | 1.04 |
ENSMUST00000238718.2
ENSMUST00000033583.14 ENSMUST00000151689.9 |
Magt1
|
magnesium transporter 1 |
| chr3_-_51184730 | 1.02 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr3_-_65299967 | 1.02 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr11_+_72498029 | 1.00 |
ENSMUST00000021148.13
ENSMUST00000138247.8 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr11_-_100713348 | 0.99 |
ENSMUST00000107358.9
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr5_-_125418107 | 0.99 |
ENSMUST00000111390.8
ENSMUST00000086075.13 |
Scarb1
|
scavenger receptor class B, member 1 |
| chr3_+_121243395 | 0.99 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr2_+_125701054 | 0.98 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chr8_-_45747883 | 0.97 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr19_-_32080496 | 0.97 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr12_-_25147139 | 0.96 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chrM_+_9459 | 0.96 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr1_+_58752415 | 0.95 |
ENSMUST00000114309.8
ENSMUST00000069333.8 |
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_85947806 | 0.95 |
ENSMUST00000238222.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr2_-_52566583 | 0.93 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_+_92831150 | 0.93 |
ENSMUST00000113055.9
|
Shroom3
|
shroom family member 3 |
| chr15_+_87509413 | 0.91 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr11_+_94218810 | 0.91 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr1_-_82564501 | 0.89 |
ENSMUST00000087050.7
|
Col4a4
|
collagen, type IV, alpha 4 |
| chrX_-_72140623 | 0.89 |
ENSMUST00000114524.9
ENSMUST00000074619.6 |
Xlr3a
|
X-linked lymphocyte-regulated 3A |
| chr5_-_38649291 | 0.89 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr6_-_34965339 | 0.88 |
ENSMUST00000201355.4
|
Slc23a4
|
solute carrier family 23 member 4 |
| chr14_+_48683581 | 0.86 |
ENSMUST00000227440.2
ENSMUST00000124720.8 ENSMUST00000226422.2 ENSMUST00000226400.2 |
Tmem260
|
transmembrane protein 260 |
| chrM_-_14061 | 0.85 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_-_57010921 | 0.82 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr6_-_52185674 | 0.82 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr3_+_144824644 | 0.81 |
ENSMUST00000199124.5
|
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr5_+_87814058 | 0.81 |
ENSMUST00000199506.5
ENSMUST00000197631.5 ENSMUST00000094641.9 |
Csn1s1
|
casein alpha s1 |
| chr3_+_151143557 | 0.81 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr1_-_175319842 | 0.80 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr14_-_59602882 | 0.80 |
ENSMUST00000160425.8
ENSMUST00000095157.11 |
Phf11d
|
PHD finger protein 11D |
| chr6_-_138013901 | 0.80 |
ENSMUST00000150278.3
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr16_-_65359566 | 0.79 |
ENSMUST00000004965.8
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr16_-_62607105 | 0.78 |
ENSMUST00000152553.2
ENSMUST00000063089.12 |
Nsun3
|
NOL1/NOP2/Sun domain family member 3 |
| chr9_-_45866550 | 0.78 |
ENSMUST00000038488.17
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr13_+_93440572 | 0.77 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr4_+_102278715 | 0.77 |
ENSMUST00000106904.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr17_+_24645615 | 0.76 |
ENSMUST00000234304.2
ENSMUST00000024946.7 ENSMUST00000234150.2 |
Eci1
|
enoyl-Coenzyme A delta isomerase 1 |
| chr6_-_90758954 | 0.75 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr7_+_143838191 | 0.73 |
ENSMUST00000097929.4
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr13_-_48666870 | 0.73 |
ENSMUST00000177530.8
ENSMUST00000176996.8 ENSMUST00000176949.2 ENSMUST00000176176.8 |
Zfp169
|
zinc finger protein 169 |
| chr9_+_110709353 | 0.72 |
ENSMUST00000155014.2
|
Als2cl
|
ALS2 C-terminal like |
| chr5_+_31684331 | 0.71 |
ENSMUST00000114533.9
ENSMUST00000202214.4 ENSMUST00000201858.4 ENSMUST00000202950.4 |
Slc4a1ap
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr15_+_39609320 | 0.68 |
ENSMUST00000227368.2
ENSMUST00000228556.2 ENSMUST00000022913.6 ENSMUST00000228701.2 ENSMUST00000227792.2 |
Dcstamp
|
dendrocyte expressed seven transmembrane protein |
| chr13_+_24986003 | 0.68 |
ENSMUST00000155575.2
|
BC005537
|
cDNA sequence BC005537 |
| chr15_+_92495007 | 0.68 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr12_+_117652526 | 0.67 |
ENSMUST00000222185.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr14_-_52728503 | 0.66 |
ENSMUST00000073571.6
|
Olfr1507
|
olfactory receptor 1507 |
| chr1_-_63215952 | 0.66 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr2_-_80277969 | 0.66 |
ENSMUST00000028389.4
|
Frzb
|
frizzled-related protein |
| chr2_-_38604503 | 0.65 |
ENSMUST00000028084.5
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr7_+_97569156 | 0.65 |
ENSMUST00000041860.13
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr5_+_93045837 | 0.64 |
ENSMUST00000113051.9
|
Shroom3
|
shroom family member 3 |
| chr15_-_36496880 | 0.64 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr3_+_132335704 | 0.63 |
ENSMUST00000212594.2
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr6_+_116627635 | 0.62 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chr17_+_85265420 | 0.61 |
ENSMUST00000080217.14
ENSMUST00000112304.10 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chrX_-_111315519 | 0.61 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr11_-_54140462 | 0.60 |
ENSMUST00000019060.6
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr6_+_21215472 | 0.60 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr5_+_92831468 | 0.60 |
ENSMUST00000168878.8
|
Shroom3
|
shroom family member 3 |
| chr14_+_51245368 | 0.60 |
ENSMUST00000022424.8
|
Rnase10
|
ribonuclease, RNase A family, 10 (non-active) |
| chr11_-_86561980 | 0.60 |
ENSMUST00000143991.3
|
Vmp1
|
vacuole membrane protein 1 |
| chr16_+_13176238 | 0.59 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr2_+_152873772 | 0.59 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr8_-_84059048 | 0.59 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr3_+_93301003 | 0.59 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chr9_+_121539395 | 0.59 |
ENSMUST00000035113.11
ENSMUST00000215966.2 ENSMUST00000215833.2 ENSMUST00000215104.2 |
Ss18l2
|
SS18, nBAF chromatin remodeling complex subunit like 2 |
| chr11_-_21951605 | 0.59 |
ENSMUST00000006071.14
|
Otx1
|
orthodenticle homeobox 1 |
| chr4_+_116453927 | 0.58 |
ENSMUST00000051869.8
|
Ccdc17
|
coiled-coil domain containing 17 |
| chr18_-_66155651 | 0.57 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr13_+_19374502 | 0.57 |
ENSMUST00000198330.2
ENSMUST00000103555.3 |
Trgv6
|
T cell receptor gamma, variable 6 |
| chr1_-_57011595 | 0.56 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr3_+_28946760 | 0.56 |
ENSMUST00000099170.2
|
Gm1527
|
predicted gene 1527 |
| chr8_+_127898139 | 0.56 |
ENSMUST00000238870.2
|
Pard3
|
par-3 family cell polarity regulator |
| chr8_+_96442509 | 0.56 |
ENSMUST00000034096.6
|
Setd6
|
SET domain containing 6 |
| chr8_-_111629074 | 0.56 |
ENSMUST00000041382.7
|
Fcsk
|
fucose kinase |
| chr16_-_65359406 | 0.55 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr6_-_122259768 | 0.53 |
ENSMUST00000032207.9
|
Klrg1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_-_57008986 | 0.53 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.7 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 2.4 | 7.3 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 2.4 | 7.3 | GO:0097037 | heme export(GO:0097037) |
| 2.4 | 7.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.4 | 15.9 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.4 | 4.2 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 1.1 | 3.4 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.9 | 9.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.9 | 4.4 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.7 | 2.7 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.7 | 2.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) positive regulation of thymocyte migration(GO:2000412) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.6 | 2.3 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.5 | 1.6 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.5 | 1.6 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.5 | 2.1 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.4 | 11.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 4.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 25.9 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.4 | 1.5 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.4 | 3.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 3.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 1.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.3 | 1.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.3 | 5.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.6 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.3 | 2.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 0.8 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.2 | 3.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.0 | GO:1901740 | skeletal muscle atrophy(GO:0014732) negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 4.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 2.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 14.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 3.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 5.7 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.2 | 1.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 1.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 0.7 | GO:0009305 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.2 | 1.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.2 | 2.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 1.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 3.0 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.7 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.2 | 1.2 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.2 | 1.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 0.5 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 5.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 0.6 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 24.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 1.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.7 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 | 0.6 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 1.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 2.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.7 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.6 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 1.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 2.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.4 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.3 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.3 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 2.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.6 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 1.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 0.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.6 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.2 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 2.6 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 3.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 1.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 2.9 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 2.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0099543 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 1.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 2.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 1.4 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 1.0 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.4 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 1.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.2 | 11.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.7 | 5.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 21.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.4 | 16.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.4 | 14.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 3.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 1.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.3 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 4.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 1.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 3.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 15.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 2.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.6 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 2.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 5.0 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 33.1 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.3 | GO:0043197 | dendritic spine(GO:0043197) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 19.7 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 1.6 | 25.9 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.5 | 9.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.2 | 25.9 | GO:0070513 | death domain binding(GO:0070513) |
| 1.2 | 7.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.9 | 7.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.9 | 3.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.8 | 33.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.7 | 3.0 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.7 | 2.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.6 | 3.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.6 | 4.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.5 | 2.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 2.1 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.4 | 4.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 2.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 1.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.3 | 4.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 1.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 14.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 7.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 2.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 1.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 5.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 7.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 4.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 1.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 2.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 0.7 | GO:0004078 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.2 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 5.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 0.8 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 5.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.8 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 1.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 3.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 5.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.8 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.8 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 3.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 10.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 3.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 2.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 8.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 1.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 5.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 17.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 12.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 14.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 7.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 5.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 8.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 2.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 2.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 5.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 4.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 4.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 3.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 2.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 7.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 2.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |