Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxc10
Z-value: 0.77
Transcription factors associated with Hoxc10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc10
|
ENSMUSG00000022484.8 | Hoxc10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc10 | mm39_v1_chr15_+_102875229_102875248 | 0.05 | 7.5e-01 | Click! |
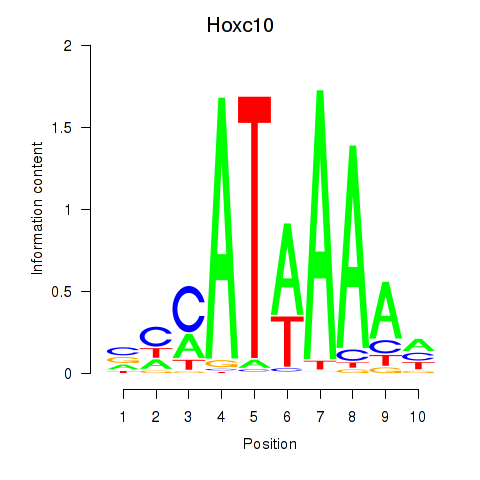
Activity profile of Hoxc10 motif
Sorted Z-values of Hoxc10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_30623592 | 2.16 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr1_-_45542442 | 1.91 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr5_+_90708962 | 1.71 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr14_-_47426863 | 1.49 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr2_-_32341408 | 1.47 |
ENSMUST00000028160.15
ENSMUST00000113310.9 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr17_-_31363245 | 1.41 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr4_+_118217179 | 1.26 |
ENSMUST00000006562.6
|
Hyi
|
hydroxypyruvate isomerase (putative) |
| chr6_-_136852792 | 1.24 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr1_-_71692320 | 1.01 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr13_+_25127127 | 0.97 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr11_-_120440519 | 0.87 |
ENSMUST00000034913.5
|
Mcrip1
|
MAPK regulated corepressor interacting protein 1 |
| chr17_+_34043536 | 0.79 |
ENSMUST00000048249.8
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chrX_-_133442596 | 0.75 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr17_+_31652073 | 0.62 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr17_+_31652029 | 0.60 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr4_-_129472328 | 0.58 |
ENSMUST00000052835.9
|
Fam167b
|
family with sequence similarity 167, member B |
| chr11_-_61268879 | 0.57 |
ENSMUST00000010267.10
|
Slc47a1
|
solute carrier family 47, member 1 |
| chr18_+_20380397 | 0.53 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr9_+_21914083 | 0.50 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr2_-_32277773 | 0.49 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr7_-_141015240 | 0.48 |
ENSMUST00000138865.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr11_-_83957889 | 0.48 |
ENSMUST00000108101.8
|
Dusp14
|
dual specificity phosphatase 14 |
| chr7_-_141014477 | 0.48 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr3_+_89173859 | 0.47 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr2_-_32278245 | 0.45 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr7_-_139162706 | 0.40 |
ENSMUST00000106095.3
|
Nkx6-2
|
NK6 homeobox 2 |
| chr11_-_73215442 | 0.39 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chrM_+_3906 | 0.37 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr11_-_100036792 | 0.36 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chrX_+_168468186 | 0.35 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr2_-_151586063 | 0.34 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr16_-_38620688 | 0.34 |
ENSMUST00000057767.6
|
Upk1b
|
uroplakin 1B |
| chr6_-_124942457 | 0.33 |
ENSMUST00000112439.9
|
Cops7a
|
COP9 signalosome subunit 7A |
| chr6_-_124942366 | 0.33 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr3_-_49711765 | 0.31 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr6_-_124942170 | 0.30 |
ENSMUST00000148485.2
ENSMUST00000129976.8 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr2_-_35994072 | 0.27 |
ENSMUST00000112961.10
ENSMUST00000112966.10 |
Lhx6
|
LIM homeobox protein 6 |
| chr3_-_49711706 | 0.26 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr17_-_34043320 | 0.24 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr3_-_88317601 | 0.24 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr15_+_6451721 | 0.24 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr5_+_43672856 | 0.24 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr8_+_94537910 | 0.24 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr17_-_34043502 | 0.23 |
ENSMUST00000087342.13
ENSMUST00000173844.8 |
Rps28
|
ribosomal protein S28 |
| chr15_-_101441254 | 0.23 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84 |
| chr1_+_189460461 | 0.23 |
ENSMUST00000097442.9
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr11_-_99134885 | 0.22 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr7_-_29605504 | 0.21 |
ENSMUST00000053521.15
|
Zfp27
|
zinc finger protein 27 |
| chr16_+_29884153 | 0.20 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr2_-_35994819 | 0.20 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr9_-_44473146 | 0.20 |
ENSMUST00000215293.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr19_+_26725589 | 0.19 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_89821818 | 0.18 |
ENSMUST00000216953.3
|
Olfr1261
|
olfactory receptor 1261 |
| chr7_-_30428930 | 0.18 |
ENSMUST00000207296.2
ENSMUST00000006478.10 |
Tmem147
|
transmembrane protein 147 |
| chr6_+_71176811 | 0.17 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr14_+_54923655 | 0.17 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr2_+_83642910 | 0.17 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chrX_+_132809189 | 0.17 |
ENSMUST00000113304.2
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr11_+_108811168 | 0.16 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr14_+_54454749 | 0.15 |
ENSMUST00000103737.3
|
Traj3
|
T cell receptor alpha joining 3 |
| chr8_-_25528972 | 0.15 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr10_-_38998272 | 0.15 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr7_-_30428746 | 0.15 |
ENSMUST00000209065.2
ENSMUST00000208169.2 |
Tmem147
|
transmembrane protein 147 |
| chr10_-_116732813 | 0.14 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr2_-_69542805 | 0.14 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr11_+_108811626 | 0.14 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr7_-_30750856 | 0.13 |
ENSMUST00000073892.6
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr3_-_63758672 | 0.12 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chr11_+_96214078 | 0.12 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr6_-_52203146 | 0.12 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr2_+_73102269 | 0.12 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr19_+_55882942 | 0.11 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr2_+_89804937 | 0.11 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr2_+_113235475 | 0.11 |
ENSMUST00000238883.2
|
Fmn1
|
formin 1 |
| chr10_+_41366433 | 0.10 |
ENSMUST00000105507.5
|
Ppil6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr2_+_177783713 | 0.10 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr15_-_101336669 | 0.10 |
ENSMUST00000081945.5
|
Krt87
|
keratin 87 |
| chr10_+_34265969 | 0.09 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chr11_+_96085118 | 0.09 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13 |
| chr2_+_3115250 | 0.09 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr10_-_33920300 | 0.09 |
ENSMUST00000048052.7
|
Calhm4
|
calcium homeostasis modulator family member 4 |
| chr8_-_62576140 | 0.09 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr7_-_103320398 | 0.08 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr5_+_149942140 | 0.08 |
ENSMUST00000065745.10
ENSMUST00000110496.5 ENSMUST00000201612.2 |
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr5_+_43673093 | 0.08 |
ENSMUST00000144558.3
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr11_+_115824108 | 0.08 |
ENSMUST00000140991.2
|
Sap30bp
|
SAP30 binding protein |
| chr11_-_99884818 | 0.07 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr9_+_37119472 | 0.07 |
ENSMUST00000034632.10
|
Tmem218
|
transmembrane protein 218 |
| chr8_-_94739469 | 0.07 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr15_+_21111428 | 0.07 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr15_-_101422054 | 0.07 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr12_-_113912416 | 0.07 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr17_-_35293447 | 0.07 |
ENSMUST00000007259.4
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr10_-_28862289 | 0.07 |
ENSMUST00000152363.8
ENSMUST00000015663.7 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr7_-_30750828 | 0.07 |
ENSMUST00000206341.2
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr10_+_90412827 | 0.06 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_+_106245792 | 0.06 |
ENSMUST00000172306.3
|
Dusp7
|
dual specificity phosphatase 7 |
| chr2_-_129213050 | 0.06 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr14_+_66043281 | 0.06 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr16_-_89305201 | 0.06 |
ENSMUST00000056118.4
|
Krtap7-1
|
keratin associated protein 7-1 |
| chr18_-_43032514 | 0.06 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_-_86180622 | 0.05 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr1_-_144124871 | 0.05 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr1_-_36283326 | 0.05 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr13_-_21765822 | 0.05 |
ENSMUST00000206526.3
ENSMUST00000213912.2 |
Olfr1364
|
olfactory receptor 1364 |
| chr14_-_50479161 | 0.05 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr5_-_67256578 | 0.04 |
ENSMUST00000012664.11
|
Phox2b
|
paired-like homeobox 2b |
| chr1_-_69147185 | 0.04 |
ENSMUST00000121473.8
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr18_-_43032359 | 0.04 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_66426127 | 0.04 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr10_-_49659355 | 0.04 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr4_-_58787716 | 0.04 |
ENSMUST00000216719.2
|
Olfr267
|
olfactory receptor 267 |
| chr11_-_99920694 | 0.04 |
ENSMUST00000073890.4
|
Krt33b
|
keratin 33B |
| chr1_-_144124787 | 0.03 |
ENSMUST00000185714.2
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr18_+_20443795 | 0.03 |
ENSMUST00000077146.4
|
Dsg1a
|
desmoglein 1 alpha |
| chr4_+_102111936 | 0.03 |
ENSMUST00000106907.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_27354006 | 0.03 |
ENSMUST00000164296.8
|
Brd3
|
bromodomain containing 3 |
| chrX_+_138464065 | 0.02 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr3_+_31956814 | 0.02 |
ENSMUST00000192429.6
ENSMUST00000191869.6 ENSMUST00000178668.2 ENSMUST00000119970.8 |
Kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chrX_-_49108236 | 0.02 |
ENSMUST00000213556.3
ENSMUST00000213463.2 |
Olfr1323
|
olfactory receptor 1323 |
| chr2_+_89821565 | 0.02 |
ENSMUST00000111509.3
ENSMUST00000213909.3 |
Olfr1261
|
olfactory receptor 1261 |
| chr11_-_115824290 | 0.01 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr3_-_117153802 | 0.01 |
ENSMUST00000197743.2
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr3_+_63988968 | 0.01 |
ENSMUST00000029406.6
|
Vmn2r1
|
vomeronasal 2, receptor 1 |
| chr8_-_18791557 | 0.01 |
ENSMUST00000033846.7
|
Angpt2
|
angiopoietin 2 |
| chr1_+_17672117 | 0.01 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chr12_-_69838004 | 0.01 |
ENSMUST00000221646.2
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr12_+_117621644 | 0.01 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr9_-_21199232 | 0.01 |
ENSMUST00000184326.8
ENSMUST00000038671.10 |
Kri1
|
KRI1 homolog |
| chr6_-_23655130 | 0.01 |
ENSMUST00000104979.2
|
Rnf148
|
ring finger protein 148 |
| chr11_-_99996452 | 0.01 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr5_+_71857261 | 0.00 |
ENSMUST00000031122.9
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr18_+_13139992 | 0.00 |
ENSMUST00000041676.3
ENSMUST00000234084.2 ENSMUST00000234565.2 |
Hrh4
|
histamine receptor H4 |
| chr15_+_92495007 | 0.00 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr14_-_50479323 | 0.00 |
ENSMUST00000214152.2
|
Olfr731
|
olfactory receptor 731 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.5 | 1.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.5 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.3 | 1.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 0.9 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 1.0 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 1.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.2 | GO:2000978 | negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 1.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 1.0 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.0 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.0 | 0.0 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 1.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.6 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.0 | 1.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |