Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Ikzf2
Z-value: 1.39
Transcription factors associated with Ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ikzf2
|
ENSMUSG00000025997.14 | Ikzf2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ikzf2 | mm39_v1_chr1_-_69724939_69724987 | 0.63 | 4.1e-05 | Click! |
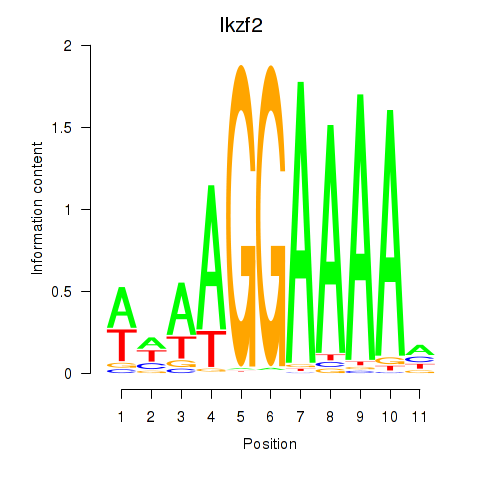
Activity profile of Ikzf2 motif
Sorted Z-values of Ikzf2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ikzf2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_103800553 | 5.63 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr15_+_80456775 | 5.53 |
ENSMUST00000229980.2
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr2_+_103800459 | 5.53 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr16_+_32427789 | 5.38 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr14_-_70864448 | 5.08 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr14_-_70864666 | 4.62 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr10_+_115653152 | 4.46 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr5_-_148329615 | 3.47 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr15_-_66703471 | 3.41 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr5_+_110478558 | 3.37 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr6_-_40562700 | 3.25 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr11_+_31822211 | 3.11 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr11_-_96859484 | 3.07 |
ENSMUST00000107623.8
|
Sp2
|
Sp2 transcription factor |
| chr16_-_75706161 | 3.06 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr11_+_31823096 | 3.02 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr15_+_79578141 | 2.93 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr17_+_73144531 | 2.84 |
ENSMUST00000233886.2
|
Ypel5
|
yippee like 5 |
| chr8_-_81466126 | 2.69 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr9_+_123902143 | 2.60 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr10_+_129219952 | 2.58 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr4_-_149783097 | 2.48 |
ENSMUST00000038859.14
ENSMUST00000105690.9 |
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chrX_+_70600481 | 2.48 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr6_+_72526236 | 2.46 |
ENSMUST00000114071.8
|
Capg
|
capping protein (actin filament), gelsolin-like |
| chr1_-_133537953 | 2.44 |
ENSMUST00000164574.2
ENSMUST00000166291.8 ENSMUST00000164096.2 ENSMUST00000166915.8 |
Snrpe
|
small nuclear ribonucleoprotein E |
| chr11_-_78074377 | 2.35 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr13_-_103901010 | 2.35 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr4_+_134195631 | 2.30 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr16_-_19801781 | 2.25 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr3_+_60436163 | 2.17 |
ENSMUST00000194201.6
ENSMUST00000193517.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr10_+_58230203 | 2.07 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr10_+_7543260 | 2.01 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr10_+_58230183 | 1.97 |
ENSMUST00000020077.11
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_+_60436570 | 1.97 |
ENSMUST00000192607.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr13_-_61084358 | 1.96 |
ENSMUST00000225859.2
ENSMUST00000225167.2 ENSMUST00000021880.10 |
Gm49391
Ctla2a
|
predicted gene, 49391 cytotoxic T lymphocyte-associated protein 2 alpha |
| chr19_-_53932581 | 1.95 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr4_-_149783009 | 1.94 |
ENSMUST00000134534.8
ENSMUST00000146612.8 ENSMUST00000105688.10 |
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr1_-_160079007 | 1.85 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr2_-_25498459 | 1.84 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr2_-_26299173 | 1.82 |
ENSMUST00000145701.8
|
Inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chrX_+_162923474 | 1.73 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr13_-_76205115 | 1.71 |
ENSMUST00000056130.8
|
Gpr150
|
G protein-coupled receptor 150 |
| chrX_+_133501928 | 1.69 |
ENSMUST00000074950.11
ENSMUST00000113203.2 ENSMUST00000113202.8 ENSMUST00000050331.13 ENSMUST00000059297.6 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr16_-_90524214 | 1.67 |
ENSMUST00000099554.5
|
Mis18a
|
MIS18 kinetochore protein A |
| chr5_+_108213608 | 1.65 |
ENSMUST00000081567.11
ENSMUST00000170319.8 ENSMUST00000112626.8 |
Mtf2
|
metal response element binding transcription factor 2 |
| chr13_+_23759930 | 1.64 |
ENSMUST00000105105.4
|
H3c4
|
H3 clustered histone 4 |
| chr7_+_46496929 | 1.61 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr4_-_92079986 | 1.57 |
ENSMUST00000123179.2
|
Gm12666
|
predicted gene 12666 |
| chr6_+_72526414 | 1.53 |
ENSMUST00000155705.8
|
Capg
|
capping protein (actin filament), gelsolin-like |
| chr19_-_53932867 | 1.46 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr2_-_72810754 | 1.46 |
ENSMUST00000066003.7
|
Sp3
|
trans-acting transcription factor 3 |
| chr1_-_84818223 | 1.43 |
ENSMUST00000186465.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr6_+_4003904 | 1.42 |
ENSMUST00000031670.10
|
Gng11
|
guanine nucleotide binding protein (G protein), gamma 11 |
| chr4_-_44168252 | 1.40 |
ENSMUST00000145760.8
ENSMUST00000128426.8 |
Rnf38
|
ring finger protein 38 |
| chrX_+_156481906 | 1.38 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr8_+_79755194 | 1.35 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr2_-_72810782 | 1.35 |
ENSMUST00000102689.10
|
Sp3
|
trans-acting transcription factor 3 |
| chr7_+_46496506 | 1.34 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr2_-_26299112 | 1.30 |
ENSMUST00000114090.8
|
Inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr5_+_31070739 | 1.29 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr12_-_111947487 | 1.21 |
ENSMUST00000190536.2
|
Rd3l
|
retinal degeneration 3-like |
| chr8_+_121215155 | 1.21 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr10_-_68377672 | 1.21 |
ENSMUST00000020103.9
|
Cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr17_-_56878438 | 1.19 |
ENSMUST00000131056.2
|
Safb2
|
scaffold attachment factor B2 |
| chr12_-_111947536 | 1.18 |
ENSMUST00000185354.2
|
Rd3l
|
retinal degeneration 3-like |
| chrX_+_156482116 | 1.18 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chrX_+_41241049 | 1.16 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr12_-_83486669 | 1.16 |
ENSMUST00000177801.2
|
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr9_-_41068771 | 1.15 |
ENSMUST00000136530.8
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chr7_+_46496552 | 1.14 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr4_-_121072264 | 1.12 |
ENSMUST00000056635.13
|
Rlf
|
rearranged L-myc fusion sequence |
| chr1_-_118239146 | 1.12 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr2_+_174171799 | 1.11 |
ENSMUST00000109085.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr13_+_23922783 | 1.09 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr3_+_89043440 | 1.07 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chrX_+_111003193 | 1.03 |
ENSMUST00000130247.9
ENSMUST00000038546.7 |
Tex16
|
testis expressed gene 16 |
| chr6_-_35110560 | 1.03 |
ENSMUST00000202143.4
ENSMUST00000114993.9 ENSMUST00000114989.9 ENSMUST00000044163.10 ENSMUST00000202417.2 |
Cnot4
|
CCR4-NOT transcription complex, subunit 4 |
| chr12_-_113823290 | 1.03 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr3_-_95902949 | 1.02 |
ENSMUST00000123006.8
ENSMUST00000130043.8 |
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr18_+_69652837 | 1.01 |
ENSMUST00000201410.4
ENSMUST00000202937.4 |
Tcf4
|
transcription factor 4 |
| chr3_-_69767208 | 0.97 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr17_+_24026892 | 0.92 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr10_-_18890281 | 0.91 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr2_+_112097087 | 0.91 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr13_-_54737857 | 0.90 |
ENSMUST00000148222.2
ENSMUST00000026987.12 |
Nop16
|
NOP16 nucleolar protein |
| chr3_-_95903313 | 0.90 |
ENSMUST00000015889.10
|
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr18_+_69652550 | 0.88 |
ENSMUST00000201205.4
|
Tcf4
|
transcription factor 4 |
| chr13_+_83652352 | 0.88 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr9_+_123901979 | 0.85 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr5_-_116162415 | 0.82 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr18_+_69652751 | 0.82 |
ENSMUST00000200966.4
|
Tcf4
|
transcription factor 4 |
| chr15_-_77037756 | 0.82 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_-_53933052 | 0.81 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr15_-_77037972 | 0.78 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_-_83033508 | 0.78 |
ENSMUST00000100375.11
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr7_-_133833734 | 0.77 |
ENSMUST00000134504.8
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr1_-_39517761 | 0.73 |
ENSMUST00000193823.2
ENSMUST00000054462.11 |
Tbc1d8
|
TBC1 domain family, member 8 |
| chr19_-_29721012 | 0.71 |
ENSMUST00000175764.9
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr3_-_95903145 | 0.71 |
ENSMUST00000143485.2
|
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr12_-_113700190 | 0.71 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr6_+_92793440 | 0.70 |
ENSMUST00000057977.4
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr5_-_103359117 | 0.69 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr15_-_83033559 | 0.69 |
ENSMUST00000058793.14
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr2_-_31006302 | 0.68 |
ENSMUST00000149196.2
|
Fnbp1
|
formin binding protein 1 |
| chr18_+_69652880 | 0.68 |
ENSMUST00000200813.4
|
Tcf4
|
transcription factor 4 |
| chr12_-_113666198 | 0.67 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr3_+_159545309 | 0.67 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chrX_+_105964224 | 0.67 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr3_+_84832783 | 0.62 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr1_-_69725045 | 0.61 |
ENSMUST00000190771.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr3_+_75982890 | 0.59 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr18_-_15284476 | 0.58 |
ENSMUST00000025992.7
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr19_-_59931432 | 0.57 |
ENSMUST00000170819.2
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr6_-_128599833 | 0.57 |
ENSMUST00000171306.5
ENSMUST00000204819.3 ENSMUST00000032512.15 |
Klrb1a
|
killer cell lectin-like receptor subfamily B member 1A |
| chr9_-_13245834 | 0.56 |
ENSMUST00000110582.4
|
Jrkl
|
Jrk-like |
| chr7_-_133833854 | 0.54 |
ENSMUST00000127524.2
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr17_-_75858835 | 0.52 |
ENSMUST00000234785.2
ENSMUST00000112507.4 |
Fam98a
|
family with sequence similarity 98, member A |
| chr1_-_69724939 | 0.51 |
ENSMUST00000027146.9
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr16_+_92089268 | 0.51 |
ENSMUST00000047383.10
|
Kcne2
|
potassium voltage-gated channel, Isk-related subfamily, gene 2 |
| chr9_+_53448322 | 0.49 |
ENSMUST00000035850.8
|
Npat
|
nuclear protein in the AT region |
| chr18_-_36859732 | 0.49 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr2_-_7400780 | 0.48 |
ENSMUST00000002176.13
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr13_+_49761506 | 0.44 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr2_+_118730823 | 0.44 |
ENSMUST00000151162.2
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chrX_-_166907286 | 0.43 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr12_+_98737274 | 0.43 |
ENSMUST00000021399.9
|
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr5_+_4073343 | 0.42 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr10_-_44334683 | 0.41 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr19_+_55882942 | 0.39 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr5_-_88675700 | 0.38 |
ENSMUST00000087033.6
|
Jchain
|
immunoglobulin joining chain |
| chr11_+_67090878 | 0.34 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr17_+_6320731 | 0.32 |
ENSMUST00000088940.6
|
Tmem181a
|
transmembrane protein 181A |
| chr3_-_61273228 | 0.31 |
ENSMUST00000066298.3
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr19_-_41065544 | 0.31 |
ENSMUST00000087176.8
|
Opalin
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr18_+_47245204 | 0.31 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr18_-_4352944 | 0.30 |
ENSMUST00000025078.10
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chrX_+_163156359 | 0.30 |
ENSMUST00000033751.8
|
Vegfd
|
vascular endothelial growth factor D |
| chr11_+_22970464 | 0.29 |
ENSMUST00000094363.4
ENSMUST00000151877.2 |
Fam161a
|
family with sequence similarity 161, member A |
| chr5_+_53747556 | 0.29 |
ENSMUST00000037618.13
ENSMUST00000201912.4 |
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr10_-_89270554 | 0.29 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr1_-_156766381 | 0.29 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr10_-_44334711 | 0.28 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr17_+_17594808 | 0.27 |
ENSMUST00000232396.2
ENSMUST00000232199.2 |
Riok2
|
RIO kinase 2 |
| chr15_-_83033471 | 0.25 |
ENSMUST00000129372.2
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr6_-_128765529 | 0.25 |
ENSMUST00000167691.9
ENSMUST00000174404.8 |
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr5_+_20433169 | 0.24 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr10_+_107998033 | 0.23 |
ENSMUST00000219263.2
|
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr5_-_51725059 | 0.23 |
ENSMUST00000127135.3
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr15_-_77037926 | 0.22 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrX_-_87159237 | 0.22 |
ENSMUST00000113966.8
ENSMUST00000113964.2 |
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr3_+_32451820 | 0.22 |
ENSMUST00000108243.8
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr11_+_29413734 | 0.21 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr1_-_156766351 | 0.18 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr9_-_117081518 | 0.18 |
ENSMUST00000111773.10
ENSMUST00000068962.14 ENSMUST00000044901.14 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr12_+_76371679 | 0.18 |
ENSMUST00000172992.2
|
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr11_+_69909659 | 0.17 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr12_+_76371634 | 0.17 |
ENSMUST00000154078.3
ENSMUST00000095610.9 |
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr10_+_25235748 | 0.16 |
ENSMUST00000218903.2
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr2_-_7400997 | 0.14 |
ENSMUST00000137733.9
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr13_+_83652150 | 0.14 |
ENSMUST00000198199.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr6_-_121450547 | 0.13 |
ENSMUST00000046373.8
ENSMUST00000151397.3 |
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr2_-_140513320 | 0.13 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr2_+_72128239 | 0.13 |
ENSMUST00000144111.2
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chrX_+_133088238 | 0.12 |
ENSMUST00000064476.5
|
Arl13a
|
ADP-ribosylation factor-like 13A |
| chr16_+_41353360 | 0.12 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr6_-_57938488 | 0.10 |
ENSMUST00000228097.2
|
Vmn1r24
|
vomeronasal 1 receptor 24 |
| chr13_+_83652280 | 0.10 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr5_-_3707166 | 0.09 |
ENSMUST00000196304.2
|
Gatad1
|
GATA zinc finger domain containing 1 |
| chr5_+_108213562 | 0.07 |
ENSMUST00000172045.8
|
Mtf2
|
metal response element binding transcription factor 2 |
| chr7_+_91321500 | 0.07 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr13_-_57043550 | 0.06 |
ENSMUST00000022023.13
ENSMUST00000174457.9 ENSMUST00000109871.8 |
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr2_-_140513382 | 0.06 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr16_-_26810402 | 0.05 |
ENSMUST00000231299.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr6_-_119173699 | 0.04 |
ENSMUST00000239204.2
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr14_-_72840373 | 0.04 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr16_-_88571089 | 0.04 |
ENSMUST00000054223.4
|
2310057N15Rik
|
RIKEN cDNA 2310057N15 gene |
| chr2_+_57887896 | 0.02 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chrX_-_4194587 | 0.01 |
ENSMUST00000179325.2
|
Btbd35f20
|
BTB domain containing 35, family member 20 |
| chr3_-_37180093 | 0.01 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr11_-_55310724 | 0.01 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr1_+_45350698 | 0.00 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chrX_+_102465616 | 0.00 |
ENSMUST00000182089.2
|
Gm26992
|
predicted gene, 26992 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.2 | 3.5 | GO:2000451 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 1.1 | 3.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.7 | 2.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.7 | 11.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.6 | 6.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 4.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 1.7 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.4 | 2.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.4 | 4.1 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 | 4.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 1.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.3 | 5.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.3 | 2.3 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 0.9 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.3 | 1.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 2.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 4.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 1.4 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.2 | 0.7 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.2 | 0.9 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 0.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 3.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 1.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 1.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 3.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 2.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.5 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 3.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.5 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 1.0 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 3.3 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.1 | 1.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 4.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 3.1 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 4.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 5.6 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 3.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 2.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 2.0 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.8 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.2 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 1.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.1 | 2.9 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 1.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 2.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.4 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) |
| 0.0 | 0.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 1.6 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:2000197 | regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 1.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 2.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 2.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 2.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 1.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 3.1 | GO:0035264 | multicellular organism growth(GO:0035264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.7 | 3.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 5.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 2.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.3 | 1.0 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.3 | 4.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 2.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 3.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 6.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 4.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 2.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 2.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 4.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 2.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 2.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 4.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.0 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.2 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 4.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 2.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 2.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 2.2 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 10.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.0 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.0 | 1.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 5.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 1.1 | 5.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 1.0 | 3.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.4 | 1.3 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.4 | 3.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 4.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 4.6 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.3 | 3.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 2.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 6.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 4.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 1.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.9 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 1.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 3.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 2.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 11.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 9.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 1.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 3.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 1.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 2.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 4.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 9.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 4.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 3.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 3.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 4.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 2.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 3.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 4.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 5.8 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 4.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 4.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 4.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |