Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Irf5_Irf6
Z-value: 0.85
Transcription factors associated with Irf5_Irf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
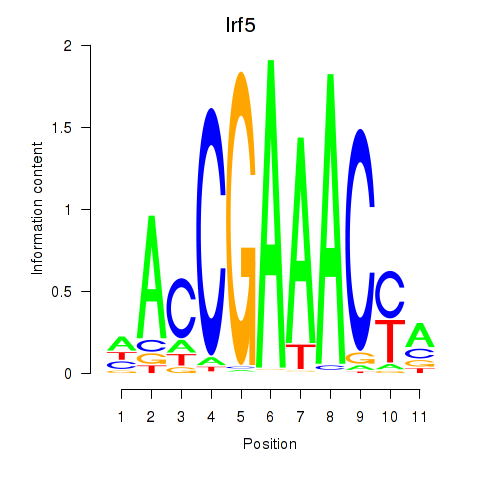
Irf5
|
ENSMUSG00000029771.13 | Irf5 |
|
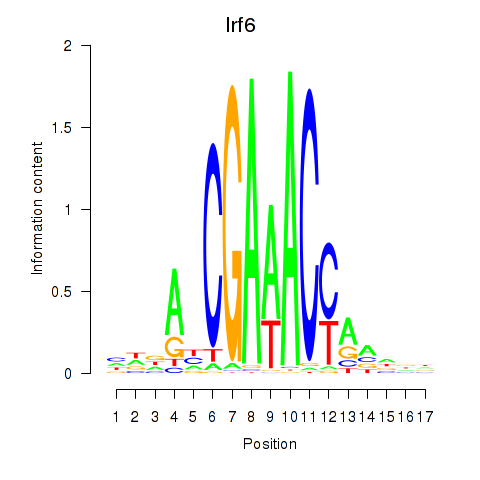
Irf6
|
ENSMUSG00000026638.16 | Irf6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf6 | mm39_v1_chr1_+_192835414_192835516 | 0.54 | 6.2e-04 | Click! |
| Irf5 | mm39_v1_chr6_+_29531247_29531269 | 0.11 | 5.2e-01 | Click! |
Activity profile of Irf5_Irf6 motif
Sorted Z-values of Irf5_Irf6 motif
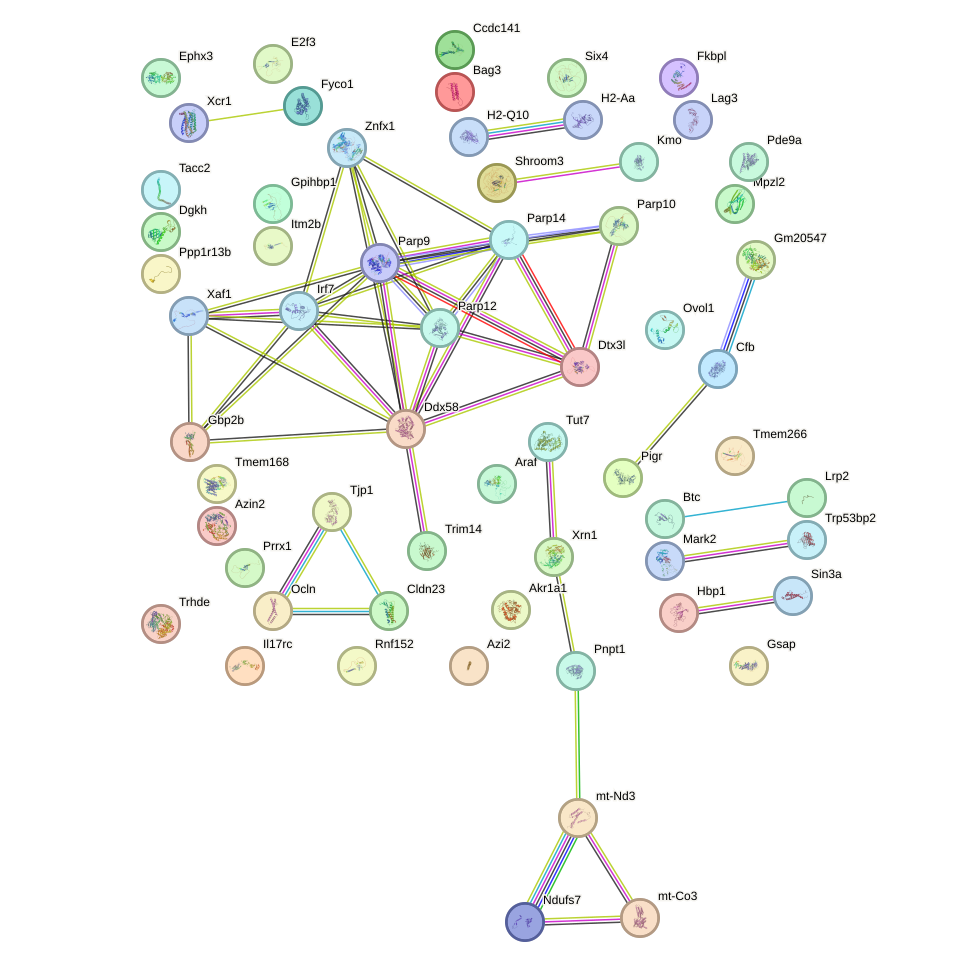
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf5_Irf6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_35081129 | 4.30 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr17_-_35081456 | 4.25 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr1_+_130754413 | 3.65 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr5_+_21391282 | 1.88 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr17_+_31652073 | 1.84 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr17_+_31652029 | 1.78 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr3_-_142101418 | 1.73 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr9_+_44953723 | 1.48 |
ENSMUST00000034600.5
|
Mpzl2
|
myelin protein zero-like 2 |
| chr17_+_35780977 | 1.46 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr4_-_46536088 | 1.39 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr6_-_39095144 | 1.36 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr14_-_73622638 | 1.26 |
ENSMUST00000228637.2
ENSMUST00000022704.9 |
Itm2b
|
integral membrane protein 2B |
| chr4_-_40239778 | 1.25 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr16_-_35691914 | 1.24 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr9_-_123680927 | 1.24 |
ENSMUST00000184082.3
ENSMUST00000167595.9 |
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr1_+_52158693 | 1.21 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr6_+_113448388 | 1.12 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr1_+_52158599 | 1.11 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_+_52158721 | 1.07 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chrM_+_9459 | 1.06 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr9_-_123680726 | 1.05 |
ENSMUST00000084715.14
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr14_-_78970160 | 1.05 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr9_+_55234197 | 0.97 |
ENSMUST00000085754.10
ENSMUST00000034862.5 |
Tmem266
|
transmembrane protein 266 |
| chr6_-_13607963 | 0.96 |
ENSMUST00000031554.9
ENSMUST00000149123.3 |
Tmem168
|
transmembrane protein 168 |
| chr1_+_175459559 | 0.91 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_+_175459735 | 0.89 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr17_-_34506744 | 0.89 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr3_-_142101339 | 0.87 |
ENSMUST00000198381.5
ENSMUST00000090134.12 ENSMUST00000196908.5 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr7_+_128125339 | 0.84 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chr13_-_100689212 | 0.82 |
ENSMUST00000022140.12
|
Ocln
|
occludin |
| chr7_-_140846328 | 0.78 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr6_-_84564623 | 0.75 |
ENSMUST00000205228.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr13_-_100688949 | 0.73 |
ENSMUST00000159515.2
ENSMUST00000160859.8 ENSMUST00000069756.11 |
Ocln
|
occludin |
| chr16_+_23428650 | 0.72 |
ENSMUST00000038423.6
ENSMUST00000211349.2 |
Rtp4
|
receptor transporter protein 4 |
| chr7_-_65020655 | 0.69 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr10_+_80085275 | 0.68 |
ENSMUST00000020361.7
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr11_-_78875657 | 0.67 |
ENSMUST00000073001.5
|
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr16_+_35758836 | 0.61 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr11_-_78875689 | 0.58 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr13_-_100689105 | 0.58 |
ENSMUST00000159459.8
|
Ocln
|
occludin |
| chr12_-_32000209 | 0.55 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr5_-_91550853 | 0.55 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr2_-_6327884 | 0.55 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene |
| chr11_+_72192455 | 0.52 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr6_-_124888643 | 0.52 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr2_-_69416365 | 0.51 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr2_-_77000878 | 0.49 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr12_-_32000169 | 0.48 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr6_-_38331187 | 0.46 |
ENSMUST00000114900.8
ENSMUST00000143702.5 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr4_-_40239700 | 0.45 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr1_-_105284383 | 0.45 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr16_-_67417687 | 0.44 |
ENSMUST00000120594.8
|
Cadm2
|
cell adhesion molecule 2 |
| chr16_-_35759461 | 0.42 |
ENSMUST00000081933.14
ENSMUST00000114885.3 |
Dtx3l
|
deltex 3-like, E3 ubiquitin ligase |
| chrM_+_8603 | 0.42 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr4_-_116508842 | 0.36 |
ENSMUST00000030455.15
|
Akr1a1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr1_+_182236728 | 0.35 |
ENSMUST00000117245.2
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chr16_-_67417768 | 0.35 |
ENSMUST00000114292.8
ENSMUST00000120898.8 |
Cadm2
|
cell adhesion molecule 2 |
| chr2_-_166904902 | 0.32 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr1_-_105284407 | 0.30 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr2_-_77349909 | 0.30 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr15_-_76127600 | 0.28 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr1_-_75293997 | 0.27 |
ENSMUST00000189282.3
ENSMUST00000191254.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr7_+_130294403 | 0.23 |
ENSMUST00000207282.2
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr9_+_95836839 | 0.22 |
ENSMUST00000189106.2
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr12_-_73160181 | 0.22 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr8_-_36293588 | 0.21 |
ENSMUST00000060128.7
|
Cldn23
|
claudin 23 |
| chr3_+_142202642 | 0.20 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chrX_+_20714782 | 0.20 |
ENSMUST00000001155.11
ENSMUST00000122312.8 ENSMUST00000120356.8 ENSMUST00000122850.2 |
Araf
|
Araf proto-oncogene, serine/threonine kinase |
| chr13_-_30170031 | 0.19 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr9_+_95836869 | 0.19 |
ENSMUST00000190665.2
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr1_-_163141278 | 0.19 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr9_+_117869642 | 0.19 |
ENSMUST00000134433.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr19_-_7272758 | 0.18 |
ENSMUST00000025921.15
|
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr9_+_117869577 | 0.18 |
ENSMUST00000133580.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr6_-_70313491 | 0.17 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr17_-_32408431 | 0.17 |
ENSMUST00000087721.10
ENSMUST00000162117.3 |
Ephx3
|
epoxide hydrolase 3 |
| chr2_-_77000936 | 0.17 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr19_+_44744484 | 0.16 |
ENSMUST00000174490.9
|
Pax2
|
paired box 2 |
| chr1_-_163141230 | 0.16 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chr9_+_117869543 | 0.15 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr1_-_75293942 | 0.15 |
ENSMUST00000187075.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr12_+_119354110 | 0.14 |
ENSMUST00000222058.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr5_+_92957231 | 0.13 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3 |
| chr11_+_29080733 | 0.13 |
ENSMUST00000020756.9
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr10_-_114638202 | 0.13 |
ENSMUST00000239411.2
|
Trhde
|
TRH-degrading enzyme |
| chr13_-_59970383 | 0.11 |
ENSMUST00000225987.2
|
Tut7
|
terminal uridylyl transferase 7 |
| chr17_+_34863738 | 0.09 |
ENSMUST00000036720.9
|
Fkbpl
|
FK506 binding protein-like |
| chr6_+_70663930 | 0.09 |
ENSMUST00000103402.2
|
Igkv3-3
|
immunoglobulin kappa variable 3-3 |
| chr17_+_34863779 | 0.09 |
ENSMUST00000174796.2
|
Fkbpl
|
FK506 binding protein-like |
| chr12_-_32000534 | 0.07 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr8_+_62381115 | 0.07 |
ENSMUST00000154398.8
ENSMUST00000156980.8 ENSMUST00000093485.3 ENSMUST00000070631.15 |
Ddx60
|
DExD/H box helicase 60 |
| chr19_-_5610628 | 0.04 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr9_-_123691077 | 0.04 |
ENSMUST00000182350.3
|
Xcr1
|
chemokine (C motif) receptor 1 |
| chr3_+_135144481 | 0.02 |
ENSMUST00000199582.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_44912927 | 0.00 |
ENSMUST00000202432.4
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr9_+_56979307 | 0.00 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.1 | 3.4 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.5 | 8.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 2.3 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.4 | 1.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 | 1.7 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 1.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 1.3 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.2 | 3.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 1.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.8 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.8 | GO:1902811 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 2.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 1.9 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.4 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 1.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.1 | 1.1 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.5 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 1.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 1.0 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.6 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.9 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.8 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 2.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.4 | GO:0090503 | histone mRNA catabolic process(GO:0071044) RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.5 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 1.1 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 3.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 6.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 3.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 2.0 | GO:0043296 | apical junction complex(GO:0043296) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 3.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 0.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 1.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 3.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 3.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 2.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 8.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 8.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 3.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |