Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
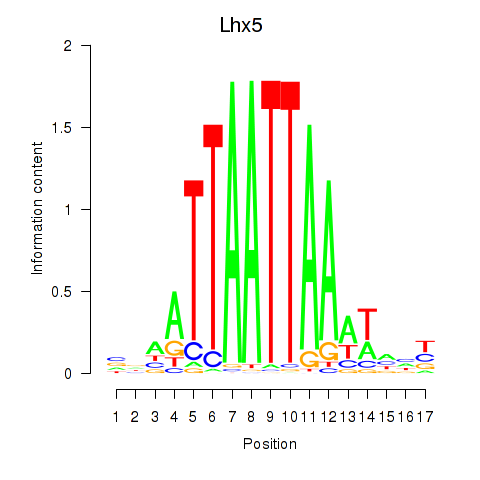
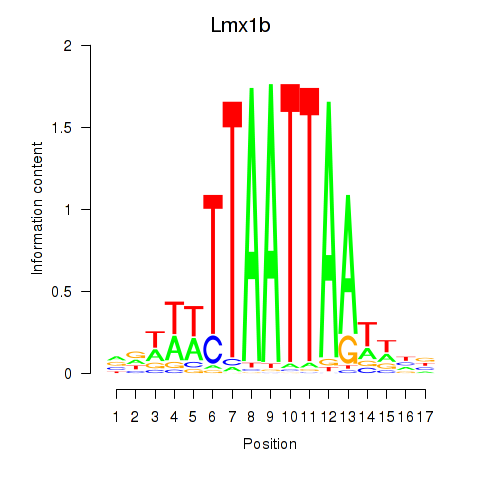
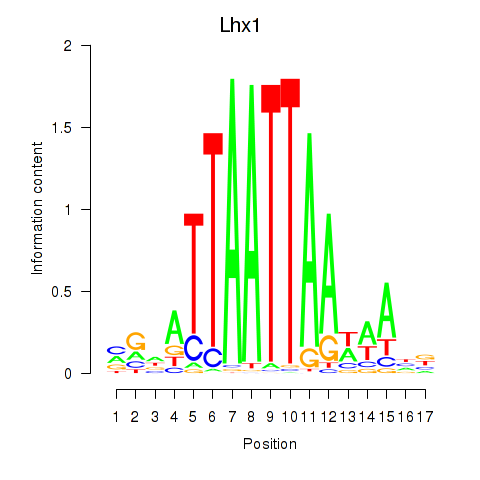
Results for Lhx5_Lmx1b_Lhx1
Z-value: 0.47
Transcription factors associated with Lhx5_Lmx1b_Lhx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx5
|
ENSMUSG00000029595.10 | Lhx5 |
|
Lmx1b
|
ENSMUSG00000038765.15 | Lmx1b |
|
Lhx1
|
ENSMUSG00000018698.16 | Lhx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx5 | mm39_v1_chr5_+_120569764_120569764 | 0.29 | 8.8e-02 | Click! |
| Lmx1b | mm39_v1_chr2_-_33530492_33530620 | 0.08 | 6.5e-01 | Click! |
| Lhx1 | mm39_v1_chr11_-_84416340_84416369 | 0.04 | 8.3e-01 | Click! |
Activity profile of Lhx5_Lmx1b_Lhx1 motif
Sorted Z-values of Lhx5_Lmx1b_Lhx1 motif
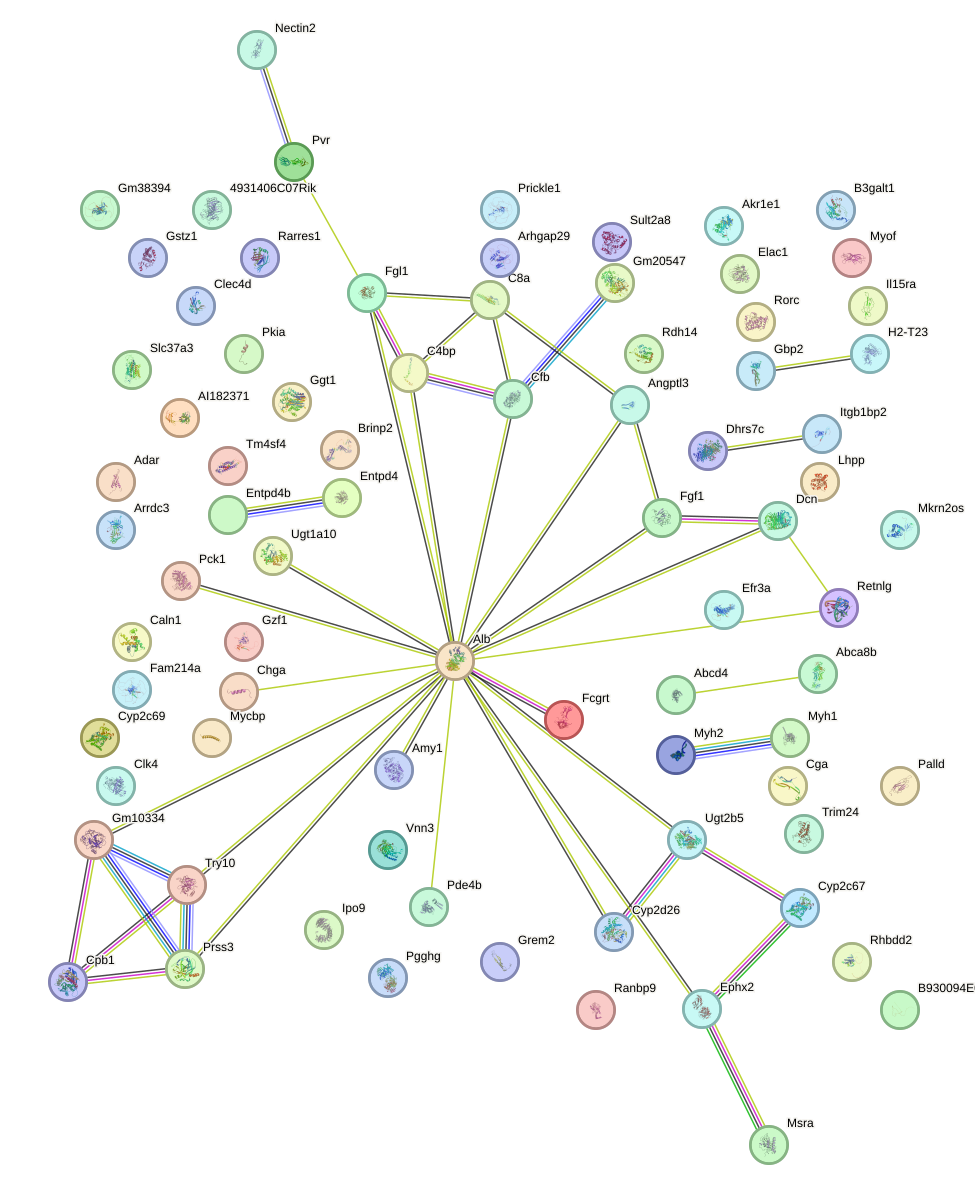
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx5_Lmx1b_Lhx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41354538 | 3.79 |
ENSMUST00000096003.7
|
Prss3
|
protease, serine 3 |
| chr6_-_41423004 | 2.66 |
ENSMUST00000095999.7
|
Gm10334
|
predicted gene 10334 |
| chr19_-_39875192 | 2.64 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr3_-_20329823 | 1.64 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr6_+_41331039 | 1.36 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr19_-_39637489 | 1.31 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr19_-_39801188 | 1.30 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr8_-_41668182 | 1.14 |
ENSMUST00000034003.5
|
Fgl1
|
fibrinogen-like protein 1 |
| chr18_-_38999755 | 1.00 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr18_-_39000056 | 0.97 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr7_-_142253247 | 0.87 |
ENSMUST00000105934.8
|
Ins2
|
insulin II |
| chr3_-_113371392 | 0.80 |
ENSMUST00000067980.12
|
Amy1
|
amylase 1, salivary |
| chr7_-_44753168 | 0.79 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr1_+_87983099 | 0.77 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr2_+_172994841 | 0.72 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr16_+_22737128 | 0.67 |
ENSMUST00000170805.9
|
Fetub
|
fetuin beta |
| chr16_+_22737227 | 0.66 |
ENSMUST00000231880.2
|
Fetub
|
fetuin beta |
| chr16_+_22737050 | 0.66 |
ENSMUST00000231768.2
|
Fetub
|
fetuin beta |
| chr1_+_87983189 | 0.60 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_+_67689094 | 0.57 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr10_+_23727325 | 0.54 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr3_-_67422821 | 0.53 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr11_+_109434519 | 0.47 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr6_+_123239076 | 0.44 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr4_+_102446883 | 0.44 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr5_+_90608751 | 0.43 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr11_+_67090878 | 0.40 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr14_-_66361931 | 0.39 |
ENSMUST00000070515.2
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr17_-_35081129 | 0.39 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr6_+_37847721 | 0.38 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr8_-_62355690 | 0.38 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr17_-_35081456 | 0.37 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr2_+_11710101 | 0.37 |
ENSMUST00000138349.8
ENSMUST00000135341.8 ENSMUST00000128156.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr10_+_97318223 | 0.37 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr7_-_14180496 | 0.36 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr17_-_36343573 | 0.33 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr10_+_75402090 | 0.30 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr6_-_68713748 | 0.29 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr16_+_48692976 | 0.29 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr11_+_51152546 | 0.29 |
ENSMUST00000130641.8
|
Clk4
|
CDC like kinase 4 |
| chr6_-_115569504 | 0.29 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chrX_+_100492684 | 0.28 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr1_-_158183894 | 0.27 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr17_+_29493113 | 0.25 |
ENSMUST00000234326.2
ENSMUST00000235117.2 |
BC004004
|
cDNA sequence BC004004 |
| chr17_+_29493049 | 0.25 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004 |
| chr15_-_82678490 | 0.25 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr18_+_31742565 | 0.25 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr3_+_94280101 | 0.24 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr5_+_90708962 | 0.24 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr2_+_67935015 | 0.23 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr6_-_68609426 | 0.22 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr3_+_142326363 | 0.22 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2 |
| chr15_+_65682066 | 0.21 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr14_-_64654592 | 0.21 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr1_-_130589321 | 0.21 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr12_-_114443071 | 0.21 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr14_-_64654397 | 0.21 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr4_+_34893772 | 0.20 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr17_+_79919267 | 0.20 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr11_+_59197746 | 0.20 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr5_-_44139099 | 0.19 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr11_-_109886569 | 0.19 |
ENSMUST00000106669.3
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr17_+_29493157 | 0.19 |
ENSMUST00000234234.2
|
BC004004
|
cDNA sequence BC004004 |
| chr7_+_132212349 | 0.19 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr5_+_130398261 | 0.18 |
ENSMUST00000086029.10
|
Caln1
|
calneuron 1 |
| chr11_-_109886601 | 0.18 |
ENSMUST00000020948.15
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr14_+_69585036 | 0.18 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr12_-_84664001 | 0.18 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr7_+_140521450 | 0.18 |
ENSMUST00000164580.3
ENSMUST00000079403.11 |
Pgghg
|
protein glucosylgalactosylhydroxylysine glucosidase |
| chr5_-_44139121 | 0.17 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr1_-_130589349 | 0.17 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr1_-_133589020 | 0.17 |
ENSMUST00000193504.6
ENSMUST00000195067.2 ENSMUST00000191896.6 ENSMUST00000194668.6 ENSMUST00000195424.6 ENSMUST00000179598.4 ENSMUST00000027736.13 |
Zc3h11a
Gm38394
Zc3h11a
|
zinc finger CCCH type containing 11A predicted gene, 38394 zinc finger CCCH type containing 11A |
| chr5_+_135661472 | 0.17 |
ENSMUST00000043707.7
|
Rhbdd2
|
rhomboid domain containing 2 |
| chr2_-_5068807 | 0.16 |
ENSMUST00000114996.8
|
Optn
|
optineurin |
| chr7_-_142209755 | 0.16 |
ENSMUST00000178921.2
|
Igf2
|
insulin-like growth factor 2 |
| chr2_-_34990689 | 0.16 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr3_+_121746862 | 0.16 |
ENSMUST00000037958.14
ENSMUST00000196904.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr7_-_15781838 | 0.15 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr19_-_24178000 | 0.15 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr1_-_171854818 | 0.15 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr9_-_15212745 | 0.15 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr13_-_43634695 | 0.15 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr1_-_174749379 | 0.15 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr3_+_89622323 | 0.14 |
ENSMUST00000098924.9
|
Adar
|
adenosine deaminase, RNA-specific |
| chr14_+_65504067 | 0.14 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr9_+_74883377 | 0.14 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr14_-_104081119 | 0.14 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr9_-_15212849 | 0.14 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr18_-_73887528 | 0.14 |
ENSMUST00000041138.3
|
Elac1
|
elaC ribonuclease Z 1 |
| chr18_+_36661198 | 0.13 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr4_+_98919183 | 0.13 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr3_+_57332735 | 0.13 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr19_-_38032006 | 0.13 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr4_-_104733580 | 0.13 |
ENSMUST00000064873.9
ENSMUST00000106808.10 ENSMUST00000048947.15 |
C8a
|
complement component 8, alpha polypeptide |
| chr2_-_5068743 | 0.12 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chr6_-_69282389 | 0.12 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr5_-_87288177 | 0.12 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr12_+_87204374 | 0.12 |
ENSMUST00000222222.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr2_+_148523118 | 0.11 |
ENSMUST00000131292.2
|
Gzf1
|
GDNF-inducible zinc finger protein 1 |
| chr15_-_93493758 | 0.11 |
ENSMUST00000048982.11
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr12_-_114710326 | 0.11 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr15_+_55171138 | 0.11 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr12_-_114487525 | 0.11 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr13_+_81034214 | 0.11 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr7_-_19655063 | 0.10 |
ENSMUST00000043517.8
|
Pvr
|
poliovirus receptor |
| chr13_-_4659120 | 0.10 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr19_-_38031774 | 0.10 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr3_+_7494108 | 0.10 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr12_+_10440755 | 0.10 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr6_-_39354570 | 0.09 |
ENSMUST00000200771.4
ENSMUST00000202204.4 ENSMUST00000202952.2 ENSMUST00000202400.4 ENSMUST00000200969.4 ENSMUST00000202749.4 ENSMUST00000090243.8 |
Slc37a3
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 3 |
| chr16_-_22676264 | 0.09 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr14_-_31139617 | 0.09 |
ENSMUST00000100730.10
|
Sh3bp5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr14_-_104081827 | 0.09 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr9_+_65538352 | 0.09 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr9_+_80072361 | 0.09 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr4_+_95445731 | 0.09 |
ENSMUST00000079223.11
ENSMUST00000177394.8 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr15_+_98468885 | 0.09 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr8_-_49008305 | 0.09 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr9_+_92339422 | 0.08 |
ENSMUST00000034941.9
|
Plscr4
|
phospholipid scramblase 4 |
| chr14_-_73613385 | 0.08 |
ENSMUST00000227454.2
|
Itm2b
|
integral membrane protein 2B |
| chr6_-_68681962 | 0.08 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr9_+_19533591 | 0.08 |
ENSMUST00000215372.2
|
Zfp317
|
zinc finger protein 317 |
| chr10_+_127226180 | 0.08 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr18_+_38429792 | 0.08 |
ENSMUST00000237211.2
|
Rnf14
|
ring finger protein 14 |
| chr7_-_101486983 | 0.07 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr15_+_99192968 | 0.07 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr2_+_148522943 | 0.07 |
ENSMUST00000028928.8
|
Gzf1
|
GDNF-inducible zinc finger protein 1 |
| chr19_+_37425180 | 0.07 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr9_+_21437440 | 0.07 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr19_+_39275518 | 0.07 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr6_+_145879839 | 0.07 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr13_+_77283632 | 0.07 |
ENSMUST00000168779.3
ENSMUST00000225605.2 |
2210408I21Rik
|
RIKEN cDNA 2210408I21 gene |
| chr9_-_31123083 | 0.06 |
ENSMUST00000217641.2
ENSMUST00000072634.15 ENSMUST00000079758.9 ENSMUST00000213254.2 |
Aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr3_-_116388334 | 0.06 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr8_-_85389470 | 0.06 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr4_+_102278715 | 0.06 |
ENSMUST00000106904.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr3_+_41697046 | 0.06 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr17_+_26934617 | 0.06 |
ENSMUST00000062519.14
ENSMUST00000144221.2 ENSMUST00000142539.8 ENSMUST00000151681.2 |
Crebrf
|
CREB3 regulatory factor |
| chr19_-_34143437 | 0.06 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr2_+_83642910 | 0.06 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chrX_+_156482116 | 0.06 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr2_-_125624754 | 0.06 |
ENSMUST00000053699.13
|
Secisbp2l
|
SECIS binding protein 2-like |
| chr10_+_34173426 | 0.06 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr5_+_92957231 | 0.06 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3 |
| chr7_-_12829100 | 0.05 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr10_-_125164399 | 0.05 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr12_-_114023935 | 0.05 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr6_-_69877961 | 0.05 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr11_+_5470652 | 0.05 |
ENSMUST00000063084.16
|
Xbp1
|
X-box binding protein 1 |
| chr8_+_121842902 | 0.05 |
ENSMUST00000054691.8
|
Foxc2
|
forkhead box C2 |
| chr10_+_129072073 | 0.05 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chr14_+_51366512 | 0.05 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr7_+_30193047 | 0.05 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr13_+_33187205 | 0.04 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr15_+_100659622 | 0.04 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr12_+_55349422 | 0.04 |
ENSMUST00000021411.15
|
Prorp
|
protein only RNase P catalytic subunit |
| chr10_-_53252210 | 0.04 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr12_+_25024913 | 0.04 |
ENSMUST00000066652.7
ENSMUST00000220459.2 ENSMUST00000222941.2 |
Kidins220
|
kinase D-interacting substrate 220 |
| chr6_+_68916540 | 0.04 |
ENSMUST00000103339.2
|
Igkv13-84
|
immunoglobulin kappa chain variable 13-84 |
| chr5_-_140986312 | 0.04 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
| chr8_+_65399831 | 0.04 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr13_+_46655617 | 0.04 |
ENSMUST00000225824.2
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr2_+_87404246 | 0.04 |
ENSMUST00000213315.2
ENSMUST00000214773.2 |
Olfr1129
|
olfactory receptor 1129 |
| chr6_-_3399451 | 0.04 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr11_+_113540154 | 0.04 |
ENSMUST00000063776.14
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr3_-_62414391 | 0.04 |
ENSMUST00000029336.6
|
Dhx36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr12_+_88327391 | 0.04 |
ENSMUST00000222695.2
|
Adck1
|
aarF domain containing kinase 1 |
| chrX_+_36675081 | 0.03 |
ENSMUST00000115179.4
|
Rhox2d
|
reproductive homeobox 2D |
| chr7_-_143203348 | 0.03 |
ENSMUST00000084396.4
ENSMUST00000075588.13 ENSMUST00000146692.8 |
Tnfrsf22
|
tumor necrosis factor receptor superfamily, member 22 |
| chr7_-_143239600 | 0.03 |
ENSMUST00000208017.2
ENSMUST00000152703.2 |
Tnfrsf23
|
tumor necrosis factor receptor superfamily, member 23 |
| chr9_-_49710058 | 0.03 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr2_+_69727563 | 0.03 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_3425159 | 0.03 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr4_+_100336003 | 0.03 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr10_-_56104732 | 0.03 |
ENSMUST00000099739.5
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr5_+_104350475 | 0.03 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr12_+_88327305 | 0.03 |
ENSMUST00000101165.9
|
Adck1
|
aarF domain containing kinase 1 |
| chr9_-_54381409 | 0.03 |
ENSMUST00000127880.8
|
Dmxl2
|
Dmx-like 2 |
| chr14_+_67470884 | 0.03 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr18_+_62681982 | 0.03 |
ENSMUST00000055725.12
ENSMUST00000162365.8 |
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr6_-_129655868 | 0.02 |
ENSMUST00000118447.2
ENSMUST00000169545.8 ENSMUST00000032270.13 ENSMUST00000032271.13 |
Klrc1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr12_+_55350023 | 0.02 |
ENSMUST00000184766.8
ENSMUST00000183475.8 ENSMUST00000183654.2 |
Prorp
|
protein only RNase P catalytic subunit |
| chr1_+_88234454 | 0.02 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr5_-_118382926 | 0.02 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr18_-_15536747 | 0.02 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr3_+_89970088 | 0.02 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr5_-_86893645 | 0.02 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr18_+_37847792 | 0.02 |
ENSMUST00000192511.2
ENSMUST00000193476.2 |
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr11_+_116734104 | 0.02 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr1_+_170136372 | 0.02 |
ENSMUST00000056991.6
|
Spata46
|
spermatogenesis associated 46 |
| chrX_+_168468186 | 0.02 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr18_+_38430205 | 0.02 |
ENSMUST00000235491.2
|
Rnf14
|
ring finger protein 14 |
| chr9_-_18297277 | 0.02 |
ENSMUST00000166825.8
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr10_+_94411119 | 0.02 |
ENSMUST00000121471.8
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr4_+_147576874 | 0.02 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr17_+_33212143 | 0.02 |
ENSMUST00000087666.11
ENSMUST00000157017.2 |
Zfp952
|
zinc finger protein 952 |
| chrM_+_7006 | 0.02 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chrX_+_36712105 | 0.02 |
ENSMUST00000072167.10
ENSMUST00000184746.2 |
Rhox2e
|
reproductive homeobox 2E |
| chr2_+_32496990 | 0.02 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr9_+_22387163 | 0.02 |
ENSMUST00000128812.8
|
Bbs9
|
Bardet-Biedl syndrome 9 (human) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1990535 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.3 | 2.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 0.7 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.2 | 5.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.4 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.1 | 1.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.7 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 1.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 7.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 2.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 | 0.1 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.0 | 0.2 | GO:0046884 | luteinizing hormone secretion(GO:0032275) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.6 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.3 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.9 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.9 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 5.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 8.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.3 | GO:0070330 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |