Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
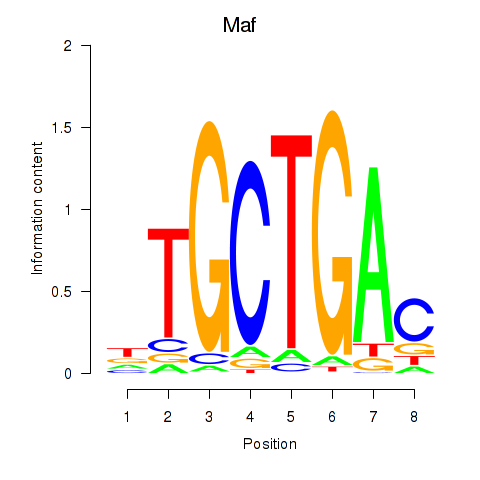
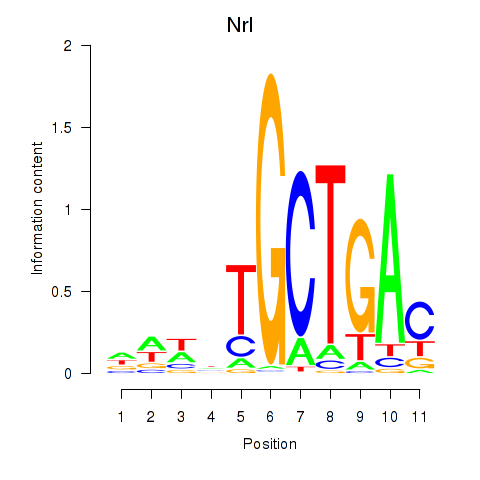
Results for Maf_Nrl
Z-value: 2.49
Transcription factors associated with Maf_Nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maf
|
ENSMUSG00000055435.7 | Maf |
|
Nrl
|
ENSMUSG00000040632.17 | Nrl |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nrl | mm39_v1_chr14_-_55762416_55762423 | -0.64 | 3.1e-05 | Click! |
| Maf | mm39_v1_chr8_-_116433733_116433835 | -0.62 | 6.3e-05 | Click! |
Activity profile of Maf_Nrl motif
Sorted Z-values of Maf_Nrl motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Maf_Nrl
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_87695352 | 24.48 |
ENSMUST00000121952.8
ENSMUST00000126490.8 |
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87695886 | 21.32 |
ENSMUST00000062862.13
|
Igf1
|
insulin-like growth factor 1 |
| chr7_+_26821266 | 19.52 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr9_-_46146558 | 15.29 |
ENSMUST00000121916.8
ENSMUST00000034586.9 |
Apoc3
|
apolipoprotein C-III |
| chr19_+_38995463 | 14.89 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr19_+_20579322 | 13.06 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr10_+_87695117 | 12.81 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr19_-_20704896 | 11.64 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr10_+_87696339 | 11.34 |
ENSMUST00000121161.8
|
Igf1
|
insulin-like growth factor 1 |
| chr4_+_104623505 | 10.59 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr10_+_87694924 | 10.36 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr9_-_46146928 | 8.77 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr16_-_45975440 | 7.31 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr5_-_77262968 | 4.92 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr4_+_140970161 | 4.64 |
ENSMUST00000138096.8
ENSMUST00000006618.9 ENSMUST00000125392.8 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr19_-_4087907 | 4.53 |
ENSMUST00000237982.2
|
Gstp1
|
glutathione S-transferase, pi 1 |
| chr19_-_4087940 | 4.48 |
ENSMUST00000237893.2
ENSMUST00000169613.4 |
Gstp1
|
glutathione S-transferase, pi 1 |
| chr1_+_127657142 | 4.10 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr19_-_4092218 | 4.09 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr15_+_10224052 | 4.06 |
ENSMUST00000128450.8
ENSMUST00000148257.8 ENSMUST00000128921.8 |
Prlr
|
prolactin receptor |
| chr19_-_21630143 | 3.36 |
ENSMUST00000179768.8
ENSMUST00000178523.2 ENSMUST00000038830.10 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr4_-_82623972 | 3.17 |
ENSMUST00000155821.2
|
Nfib
|
nuclear factor I/B |
| chr8_+_120163857 | 2.86 |
ENSMUST00000152420.8
ENSMUST00000212112.2 ENSMUST00000098365.4 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr6_+_129510331 | 2.62 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr7_+_100970435 | 2.62 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr6_+_83031502 | 2.41 |
ENSMUST00000092618.9
|
Aup1
|
ancient ubiquitous protein 1 |
| chr17_-_24428351 | 2.39 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr17_-_34250474 | 2.37 |
ENSMUST00000171872.3
ENSMUST00000025186.16 |
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr18_-_35760260 | 2.35 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr13_-_64277115 | 2.34 |
ENSMUST00000220792.2
ENSMUST00000222866.2 ENSMUST00000099441.6 ENSMUST00000222168.2 |
Slc35d2
|
solute carrier family 35, member D2 |
| chr11_-_53313950 | 2.31 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr17_-_34250616 | 2.31 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr4_-_41741278 | 2.30 |
ENSMUST00000059354.15
ENSMUST00000071561.7 |
Sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr7_+_100971034 | 2.26 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr5_-_24628514 | 2.19 |
ENSMUST00000030814.11
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr14_+_67470884 | 2.19 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr7_+_100970910 | 2.17 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr14_-_73622638 | 2.16 |
ENSMUST00000228637.2
ENSMUST00000022704.9 |
Itm2b
|
integral membrane protein 2B |
| chr2_-_32584132 | 2.14 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr16_-_56987670 | 2.12 |
ENSMUST00000023432.10
|
Nit2
|
nitrilase family, member 2 |
| chr6_-_138013901 | 2.11 |
ENSMUST00000150278.3
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr11_+_80319424 | 2.08 |
ENSMUST00000173938.8
ENSMUST00000017572.14 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr6_+_90439544 | 2.04 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr18_+_5593566 | 2.02 |
ENSMUST00000160910.2
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr18_-_43192483 | 2.00 |
ENSMUST00000025377.14
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr13_+_64309675 | 1.99 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr17_-_25394445 | 1.94 |
ENSMUST00000224277.2
|
Percc1
|
proline and glutamate rich with coiled coil 1 |
| chr2_+_34661982 | 1.93 |
ENSMUST00000028222.13
ENSMUST00000100171.3 |
Hspa5
|
heat shock protein 5 |
| chr7_+_18962252 | 1.88 |
ENSMUST00000063976.9
|
Opa3
|
optic atrophy 3 |
| chr12_+_10440755 | 1.88 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr6_+_90439596 | 1.88 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr4_-_6275629 | 1.88 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr10_-_24588030 | 1.79 |
ENSMUST00000105520.8
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr9_+_44966464 | 1.78 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr18_-_35788255 | 1.73 |
ENSMUST00000190196.5
|
Prob1
|
proline rich basic protein 1 |
| chr9_+_110074574 | 1.68 |
ENSMUST00000197850.5
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr4_+_84802592 | 1.67 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr4_-_63072367 | 1.65 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr1_-_74932266 | 1.63 |
ENSMUST00000006721.3
|
Cryba2
|
crystallin, beta A2 |
| chr3_+_95801325 | 1.61 |
ENSMUST00000197081.2
ENSMUST00000056710.10 |
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chr4_+_56740070 | 1.59 |
ENSMUST00000181745.2
|
Gm26657
|
predicted gene, 26657 |
| chr9_-_107556823 | 1.58 |
ENSMUST00000010205.9
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr3_-_33137209 | 1.57 |
ENSMUST00000194016.6
ENSMUST00000193681.6 ENSMUST00000192093.6 ENSMUST00000193289.6 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr1_-_66974694 | 1.54 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr2_+_152873772 | 1.51 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr14_+_73790105 | 1.50 |
ENSMUST00000160507.8
ENSMUST00000022706.7 |
Sucla2
|
succinate-Coenzyme A ligase, ADP-forming, beta subunit |
| chr18_+_21134928 | 1.49 |
ENSMUST00000234536.2
ENSMUST00000052396.7 |
Rnf138
|
ring finger protein 138 |
| chr3_-_58433313 | 1.49 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr15_+_57558048 | 1.48 |
ENSMUST00000096430.11
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr17_-_13271183 | 1.48 |
ENSMUST00000091648.4
|
Gpr31b
|
G protein-coupled receptor 31, D17Leh66b region |
| chr9_-_106124917 | 1.47 |
ENSMUST00000112524.9
ENSMUST00000219129.2 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr13_+_42862957 | 1.47 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr9_+_46139878 | 1.44 |
ENSMUST00000034588.9
ENSMUST00000132155.2 |
Apoa1
|
apolipoprotein A-I |
| chr2_-_156728988 | 1.44 |
ENSMUST00000029164.9
|
Sla2
|
Src-like-adaptor 2 |
| chr4_+_59003121 | 1.44 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr1_-_183150867 | 1.41 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chr2_-_156729092 | 1.41 |
ENSMUST00000109561.4
|
Sla2
|
Src-like-adaptor 2 |
| chr5_+_110692162 | 1.39 |
ENSMUST00000040001.14
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr1_+_186699613 | 1.39 |
ENSMUST00000045108.2
|
D1Pas1
|
DNA segment, Chr 1, Pasteur Institute 1 |
| chr9_-_63306497 | 1.38 |
ENSMUST00000168665.3
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chr1_-_20854490 | 1.35 |
ENSMUST00000039046.10
|
Il17f
|
interleukin 17F |
| chr3_-_33137165 | 1.35 |
ENSMUST00000078226.10
ENSMUST00000108224.8 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr18_+_21135079 | 1.33 |
ENSMUST00000234545.2
|
Rnf138
|
ring finger protein 138 |
| chr3_+_40905216 | 1.33 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr3_-_49711765 | 1.31 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr7_+_18962301 | 1.31 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chr18_-_35788240 | 1.31 |
ENSMUST00000097619.2
|
Prob1
|
proline rich basic protein 1 |
| chr17_-_26288447 | 1.30 |
ENSMUST00000122103.9
ENSMUST00000120691.9 |
Rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr14_+_54491637 | 1.29 |
ENSMUST00000180359.8
ENSMUST00000199338.2 |
Abhd4
|
abhydrolase domain containing 4 |
| chr5_-_30401416 | 1.27 |
ENSMUST00000125367.4
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr8_+_127898139 | 1.25 |
ENSMUST00000238870.2
|
Pard3
|
par-3 family cell polarity regulator |
| chr12_+_108859557 | 1.22 |
ENSMUST00000221377.2
|
Wdr25
|
WD repeat domain 25 |
| chr3_-_84128160 | 1.22 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr4_-_57300361 | 1.21 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr6_-_29380423 | 1.21 |
ENSMUST00000147483.3
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr5_-_113229445 | 1.21 |
ENSMUST00000131708.2
ENSMUST00000117143.8 ENSMUST00000119627.8 |
Crybb3
|
crystallin, beta B3 |
| chr3_+_95190255 | 1.19 |
ENSMUST00000039537.14
ENSMUST00000107187.9 |
Mindy1
|
MINDY lysine 48 deubiquitinase 1 |
| chr13_-_53627110 | 1.18 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr10_+_86541675 | 1.17 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr9_-_57513510 | 1.17 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chr10_+_4660119 | 1.15 |
ENSMUST00000105588.8
ENSMUST00000105589.2 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr10_-_23985432 | 1.14 |
ENSMUST00000041180.7
|
Taar9
|
trace amine-associated receptor 9 |
| chr11_+_69214895 | 1.14 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr12_+_80690985 | 1.13 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr2_+_6327431 | 1.12 |
ENSMUST00000114937.8
|
Usp6nl
|
USP6 N-terminal like |
| chr3_-_49711706 | 1.12 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr15_+_89338116 | 1.12 |
ENSMUST00000023291.6
|
Mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr5_-_24628483 | 1.12 |
ENSMUST00000198990.2
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr16_-_92622659 | 1.11 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr7_-_126062272 | 1.10 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr8_-_3928495 | 1.06 |
ENSMUST00000209176.2
ENSMUST00000011445.8 |
Cd209d
|
CD209d antigen |
| chr17_+_12803019 | 1.05 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr17_-_45125468 | 1.05 |
ENSMUST00000159943.8
ENSMUST00000160673.8 |
Runx2
|
runt related transcription factor 2 |
| chr2_+_125876883 | 1.04 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr5_+_31274064 | 1.04 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr11_-_100139728 | 1.03 |
ENSMUST00000007280.9
|
Krt16
|
keratin 16 |
| chr11_-_86574586 | 1.03 |
ENSMUST00000018315.10
|
Vmp1
|
vacuole membrane protein 1 |
| chr11_+_99770013 | 1.03 |
ENSMUST00000078442.4
|
Gm11567
|
predicted gene 11567 |
| chr5_-_25113358 | 1.02 |
ENSMUST00000114975.8
ENSMUST00000150135.8 |
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_+_69214883 | 1.02 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr6_+_115908709 | 1.01 |
ENSMUST00000032471.9
|
Rho
|
rhodopsin |
| chr8_-_23680954 | 1.00 |
ENSMUST00000209507.2
|
Gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr13_+_84370405 | 0.99 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chr6_+_110622533 | 0.99 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr6_-_29380467 | 0.98 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr8_-_3976808 | 0.98 |
ENSMUST00000188386.2
ENSMUST00000084086.9 ENSMUST00000171635.8 |
Cd209b
|
CD209b antigen |
| chr2_+_58991352 | 0.95 |
ENSMUST00000037903.15
|
Pkp4
|
plakophilin 4 |
| chr17_-_45125537 | 0.95 |
ENSMUST00000113571.10
|
Runx2
|
runt related transcription factor 2 |
| chr12_-_28602302 | 0.95 |
ENSMUST00000020963.8
ENSMUST00000221349.2 |
Dcdc2c
|
doublecortin domain containing 2C |
| chr5_-_77555881 | 0.93 |
ENSMUST00000163898.6
ENSMUST00000046746.10 |
Igfbp7
|
insulin-like growth factor binding protein 7 |
| chr2_+_181405106 | 0.93 |
ENSMUST00000081125.11
|
Myt1
|
myelin transcription factor 1 |
| chr16_+_20367327 | 0.92 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr5_+_31274046 | 0.92 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr1_+_85949827 | 0.91 |
ENSMUST00000131151.3
|
Spata3
|
spermatogenesis associated 3 |
| chr16_-_15681145 | 0.91 |
ENSMUST00000229680.2
|
Mzt2
|
mitotic spindle organizing protein 2 |
| chr5_+_102992873 | 0.90 |
ENSMUST00000070000.6
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr7_-_79036222 | 0.90 |
ENSMUST00000205638.2
ENSMUST00000206320.3 ENSMUST00000205442.2 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chr14_-_63402374 | 0.90 |
ENSMUST00000238526.2
|
Fdft1
|
farnesyl diphosphate farnesyl transferase 1 |
| chr15_-_85387428 | 0.90 |
ENSMUST00000178942.2
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chr7_+_98916677 | 0.89 |
ENSMUST00000127492.2
|
Map6
|
microtubule-associated protein 6 |
| chr16_+_20491465 | 0.88 |
ENSMUST00000044783.14
ENSMUST00000115463.8 ENSMUST00000142344.8 ENSMUST00000073840.12 ENSMUST00000143939.8 ENSMUST00000128594.8 ENSMUST00000150333.8 ENSMUST00000231618.2 ENSMUST00000140576.8 ENSMUST00000115457.8 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chrX_+_70860378 | 0.87 |
ENSMUST00000114575.4
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr12_+_108860192 | 0.86 |
ENSMUST00000220667.2
ENSMUST00000167816.8 ENSMUST00000047115.9 |
Wdr25
|
WD repeat domain 25 |
| chr11_+_69214789 | 0.85 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr14_-_34310637 | 0.85 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3 |
| chr7_+_27770655 | 0.85 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr1_-_65142521 | 0.84 |
ENSMUST00000061497.9
|
Cryga
|
crystallin, gamma A |
| chr16_-_62667307 | 0.82 |
ENSMUST00000232561.2
ENSMUST00000089289.6 |
Arl13b
|
ADP-ribosylation factor-like 13B |
| chr17_-_35391583 | 0.82 |
ENSMUST00000173106.2
|
Aif1
|
allograft inflammatory factor 1 |
| chr1_-_167294349 | 0.80 |
ENSMUST00000036643.6
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr11_+_69214971 | 0.79 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chrX_+_70860357 | 0.79 |
ENSMUST00000114576.9
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr2_-_26028814 | 0.79 |
ENSMUST00000163836.2
|
Tmem250-ps
|
transmembrane protein 250, pseudogene |
| chr3_+_99203818 | 0.79 |
ENSMUST00000150756.3
|
Tbx15
|
T-box 15 |
| chr9_+_7272514 | 0.79 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr16_+_17149235 | 0.78 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr3_+_45332831 | 0.77 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr13_+_83720484 | 0.77 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr3_+_135531834 | 0.76 |
ENSMUST00000029810.6
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr1_+_173877941 | 0.76 |
ENSMUST00000062665.4
|
Olfr432
|
olfactory receptor 432 |
| chr12_-_90705212 | 0.75 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr10_+_34359513 | 0.75 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr2_-_29983618 | 0.75 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr11_-_106192627 | 0.74 |
ENSMUST00000103071.4
|
Gh
|
growth hormone |
| chr14_-_34310602 | 0.74 |
ENSMUST00000064098.14
ENSMUST00000090040.12 ENSMUST00000022330.9 ENSMUST00000022327.13 |
Ldb3
|
LIM domain binding 3 |
| chr2_+_78699360 | 0.73 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr5_-_31117617 | 0.73 |
ENSMUST00000202567.2
ENSMUST00000074840.12 |
Preb
|
prolactin regulatory element binding |
| chr12_-_85871281 | 0.72 |
ENSMUST00000021676.12
ENSMUST00000142331.2 |
Erg28
|
ergosterol biosynthesis 28 |
| chr7_-_79036654 | 0.72 |
ENSMUST00000206695.2
|
Rlbp1
|
retinaldehyde binding protein 1 |
| chrX_-_103900834 | 0.72 |
ENSMUST00000033575.7
|
Magee2
|
MAGE family member E2 |
| chrX_-_90619517 | 0.72 |
ENSMUST00000078832.5
|
1700084M14Rik
|
RIKEN cDNA 1700084M14 gene |
| chr11_+_60913386 | 0.71 |
ENSMUST00000089184.11
|
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr12_+_35097179 | 0.70 |
ENSMUST00000163677.3
ENSMUST00000048519.17 |
Snx13
|
sorting nexin 13 |
| chr15_-_76084776 | 0.70 |
ENSMUST00000169108.8
ENSMUST00000170728.8 |
Plec
|
plectin |
| chr14_-_54491365 | 0.70 |
ENSMUST00000128231.2
|
Dad1
|
defender against cell death 1 |
| chr7_+_27196760 | 0.69 |
ENSMUST00000238964.2
|
Prx
|
periaxin |
| chrX_-_18327610 | 0.69 |
ENSMUST00000044188.5
|
Dipk2b
|
divergent protein kinase domain 2B |
| chr12_+_30634425 | 0.69 |
ENSMUST00000057151.10
|
Tmem18
|
transmembrane protein 18 |
| chr4_+_123010035 | 0.68 |
ENSMUST00000102648.6
|
Oxct2b
|
3-oxoacid CoA transferase 2B |
| chr14_-_34310438 | 0.68 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr3_-_64417263 | 0.67 |
ENSMUST00000177184.9
|
Vmn2r5
|
vomeronasal 2, receptor 5 |
| chr8_-_11528615 | 0.67 |
ENSMUST00000033900.7
|
Rab20
|
RAB20, member RAS oncogene family |
| chr7_+_97049210 | 0.66 |
ENSMUST00000032882.9
ENSMUST00000149122.2 |
Ndufc2
|
NADH:ubiquinone oxidoreductase subunit C2 |
| chr6_-_120470768 | 0.66 |
ENSMUST00000178687.2
|
Tmem121b
|
transmembrane protein 121B |
| chr4_+_143076327 | 0.65 |
ENSMUST00000052458.3
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr9_+_103879745 | 0.65 |
ENSMUST00000035167.15
ENSMUST00000194774.6 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr19_-_24839219 | 0.65 |
ENSMUST00000047666.5
|
Pgm5
|
phosphoglucomutase 5 |
| chr2_-_167030706 | 0.64 |
ENSMUST00000207917.2
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr19_+_38253105 | 0.64 |
ENSMUST00000196090.2
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr18_-_60866186 | 0.64 |
ENSMUST00000237885.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr8_+_122996308 | 0.64 |
ENSMUST00000055537.3
|
Zfp469
|
zinc finger protein 469 |
| chr9_-_34967081 | 0.64 |
ENSMUST00000215463.2
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr4_+_129878627 | 0.63 |
ENSMUST00000120204.8
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr11_-_77616103 | 0.63 |
ENSMUST00000078623.5
|
Cryba1
|
crystallin, beta A1 |
| chr19_+_38253077 | 0.62 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr4_-_123217391 | 0.62 |
ENSMUST00000102640.2
|
Oxct2a
|
3-oxoacid CoA transferase 2A |
| chr9_-_87613301 | 0.61 |
ENSMUST00000034991.8
|
Tbx18
|
T-box18 |
| chr6_-_56339251 | 0.61 |
ENSMUST00000166102.8
|
Pde1c
|
phosphodiesterase 1C |
| chr2_+_125876566 | 0.61 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr1_-_136188611 | 0.61 |
ENSMUST00000086395.7
|
Gpr25
|
G protein-coupled receptor 25 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.5 | 80.3 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 6.5 | 19.5 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 6.0 | 24.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 3.0 | 9.0 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 1.5 | 13.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.2 | 14.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.8 | 3.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 7.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.8 | 2.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.7 | 2.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.7 | 11.9 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.7 | 4.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 4.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.6 | 1.9 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.6 | 10.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 1.7 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.5 | 1.6 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.4 | 3.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 1.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.4 | 1.9 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.4 | 1.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.4 | 1.1 | GO:0090076 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 5.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.4 | 1.4 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.3 | 1.4 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) positive regulation of progesterone secretion(GO:2000872) |
| 0.3 | 2.8 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.3 | 3.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 1.4 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.3 | 0.8 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.3 | 2.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 1.6 | GO:0050917 | sensory perception of umami taste(GO:0050917) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.3 | 1.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.3 | 1.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.2 | 2.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 1.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 1.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.2 | 1.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 5.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 0.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.8 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 | 0.8 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.2 | 0.5 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.2 | 1.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 1.8 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 0.5 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.2 | 0.5 | GO:1901147 | optic nerve formation(GO:0021634) pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) optic chiasma development(GO:0061360) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.2 | 2.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 1.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 0.3 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 4.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 2.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 2.9 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.2 | 0.6 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 1.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 1.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 2.0 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 2.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.9 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.4 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.3 | GO:0045414 | regulation of interleukin-8 biosynthetic process(GO:0045414) |
| 0.1 | 1.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.7 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 1.1 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.1 | 2.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.4 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 3.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.7 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 1.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.6 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 0.8 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 0.9 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 2.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.6 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.7 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 2.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.8 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 1.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 2.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.2 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 1.5 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.4 | GO:0046618 | xenobiotic transport(GO:0042908) drug export(GO:0046618) |
| 0.1 | 0.3 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 0.8 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.7 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.1 | 0.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.8 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.3 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 | 0.3 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 4.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.0 | GO:0071260 | hyaluronan metabolic process(GO:0030212) cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.4 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 1.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.7 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 2.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 3.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.7 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.3 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 1.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.0 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.4 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 5.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.5 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.9 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.4 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 1.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.7 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.4 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.6 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.7 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.7 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 80.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 2.6 | 13.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 2.1 | 23.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.2 | 10.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 3.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 4.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.5 | 2.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 7.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 1.4 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.3 | 1.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 2.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 1.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 3.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 1.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 3.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.1 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.6 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 1.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 3.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 3.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 0.5 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.1 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 2.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.0 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 2.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 28.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 25.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 4.9 | 24.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 4.4 | 13.1 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 3.7 | 14.9 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.8 | 80.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.8 | 3.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.8 | 4.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.8 | 2.3 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.7 | 2.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.6 | 3.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.6 | 19.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.5 | 2.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.5 | 1.5 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.5 | 1.5 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.5 | 1.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.4 | 1.6 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 3.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.4 | 1.2 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 2.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.4 | 1.8 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 1.0 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.3 | 1.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.3 | 3.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 1.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 1.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 2.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.2 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 0.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 1.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.4 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.7 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 5.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.4 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.1 | 1.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 1.9 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.4 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 1.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.6 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 4.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 2.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 4.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 1.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.8 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.2 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 3.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.5 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.4 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 2.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 3.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 2.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 2.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 4.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 2.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 2.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0000149 | SNARE binding(GO:0000149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 79.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 3.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 3.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 13.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.2 | 23.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 2.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 3.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 10.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 4.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 5.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 9.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 6.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.7 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 3.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.8 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |