Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
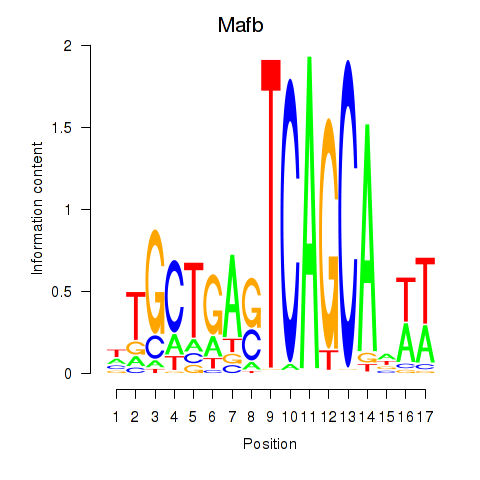
Results for Mafb
Z-value: 1.84
Transcription factors associated with Mafb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mafb
|
ENSMUSG00000074622.5 | Mafb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafb | mm39_v1_chr2_-_160208977_160208993 | -0.18 | 2.9e-01 | Click! |
Activity profile of Mafb motif
Sorted Z-values of Mafb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mafb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_96552349 | 18.41 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr7_+_140343652 | 11.41 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr1_-_130589321 | 9.55 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr17_-_12894716 | 9.53 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr1_-_130589349 | 9.46 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr8_+_105652867 | 7.52 |
ENSMUST00000034355.11
ENSMUST00000109410.4 |
Ces2e
|
carboxylesterase 2E |
| chr1_-_121255448 | 7.40 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121255400 | 6.97 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121255753 | 6.87 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr19_-_20704896 | 6.04 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr19_-_4092218 | 5.84 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr5_-_87240405 | 5.67 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr19_-_7779943 | 5.61 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr1_-_121255503 | 5.56 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr7_-_30643444 | 5.29 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr15_+_82338247 | 5.25 |
ENSMUST00000230000.2
|
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr19_-_7780025 | 5.10 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr7_+_127399776 | 4.99 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr7_+_127399789 | 4.96 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr4_+_140966810 | 4.92 |
ENSMUST00000141834.9
|
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr1_-_180027151 | 4.42 |
ENSMUST00000161743.3
|
Coq8a
|
coenzyme Q8A |
| chr3_-_131096792 | 4.31 |
ENSMUST00000200236.2
ENSMUST00000106337.7 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr1_-_51955054 | 4.19 |
ENSMUST00000018561.14
ENSMUST00000114537.9 |
Myo1b
|
myosin IB |
| chr3_+_59989282 | 4.15 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr1_-_51955126 | 4.02 |
ENSMUST00000046390.14
|
Myo1b
|
myosin IB |
| chr14_-_66361931 | 3.78 |
ENSMUST00000070515.2
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr10_+_62756409 | 3.75 |
ENSMUST00000044977.10
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr13_-_23894697 | 3.71 |
ENSMUST00000091707.13
ENSMUST00000006787.8 |
Hfe
|
homeostatic iron regulator |
| chr18_+_21094477 | 3.69 |
ENSMUST00000234316.2
|
Rnf125
|
ring finger protein 125 |
| chr15_-_100579450 | 3.67 |
ENSMUST00000230740.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr6_+_138119851 | 3.66 |
ENSMUST00000125810.2
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr6_+_128639342 | 3.66 |
ENSMUST00000032518.7
ENSMUST00000204416.2 |
Clec2h
|
C-type lectin domain family 2, member h |
| chr4_+_150203740 | 3.59 |
ENSMUST00000030826.4
|
Slc2a5
|
solute carrier family 2 (facilitated glucose transporter), member 5 |
| chr10_-_75617245 | 3.51 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr15_-_100579813 | 3.45 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chrX_+_100419965 | 3.43 |
ENSMUST00000119080.8
|
Gjb1
|
gap junction protein, beta 1 |
| chr15_+_25843225 | 3.39 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr9_-_103165489 | 3.31 |
ENSMUST00000035163.10
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr7_-_140590605 | 3.29 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr15_+_25933632 | 3.25 |
ENSMUST00000228327.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr14_+_14475188 | 3.12 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr3_-_107952146 | 3.11 |
ENSMUST00000178808.8
ENSMUST00000106670.2 ENSMUST00000029489.15 |
Gstm4
|
glutathione S-transferase, mu 4 |
| chr3_-_129126362 | 3.08 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr4_-_6275629 | 3.03 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr9_-_103165423 | 2.93 |
ENSMUST00000123530.8
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr11_-_69696428 | 2.90 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr7_+_143028831 | 2.80 |
ENSMUST00000105917.3
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr19_-_11058452 | 2.77 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr2_-_32584132 | 2.76 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr9_-_107556823 | 2.76 |
ENSMUST00000010205.9
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr4_+_138694422 | 2.61 |
ENSMUST00000116094.5
ENSMUST00000239443.2 |
Rnf186
|
ring finger protein 186 |
| chr10_-_24588030 | 2.57 |
ENSMUST00000105520.8
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr8_-_85620537 | 2.54 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr2_-_27136826 | 2.53 |
ENSMUST00000149733.8
|
Sardh
|
sarcosine dehydrogenase |
| chr11_+_48728291 | 2.52 |
ENSMUST00000046903.6
|
Trim7
|
tripartite motif-containing 7 |
| chr10_+_62756426 | 2.52 |
ENSMUST00000144459.2
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr18_+_21077627 | 2.49 |
ENSMUST00000050004.3
|
Rnf125
|
ring finger protein 125 |
| chr13_-_23894828 | 2.44 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr5_-_121523450 | 2.37 |
ENSMUST00000152265.8
|
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr15_+_82439273 | 2.32 |
ENSMUST00000229103.2
ENSMUST00000068861.8 ENSMUST00000229904.2 |
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr6_+_71470833 | 2.30 |
ENSMUST00000064637.11
ENSMUST00000114178.8 |
Rnf103
|
ring finger protein 103 |
| chr13_-_41373638 | 2.22 |
ENSMUST00000117096.2
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr12_+_17316546 | 2.20 |
ENSMUST00000057288.7
ENSMUST00000239402.2 |
Pdia6
|
protein disulfide isomerase associated 6 |
| chr3_-_89009214 | 2.18 |
ENSMUST00000081848.13
|
Fdps
|
farnesyl diphosphate synthetase |
| chr1_+_182591425 | 2.17 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr7_+_127400016 | 2.16 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr9_+_37524966 | 2.12 |
ENSMUST00000215474.2
|
Siae
|
sialic acid acetylesterase |
| chr3_-_89009153 | 2.12 |
ENSMUST00000199668.3
ENSMUST00000196709.5 |
Fdps
|
farnesyl diphosphate synthetase |
| chr5_-_121523634 | 2.10 |
ENSMUST00000120784.8
ENSMUST00000155379.8 |
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr5_-_121756928 | 2.09 |
ENSMUST00000041252.13
ENSMUST00000111776.6 |
Acad12
|
acyl-Coenzyme A dehydrogenase family, member 12 |
| chr17_-_36432041 | 2.09 |
ENSMUST00000166442.3
|
H2-T10
|
histocompatibility 2, T region locus 10 |
| chr15_-_76193955 | 2.08 |
ENSMUST00000210024.2
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr1_-_180073322 | 2.07 |
ENSMUST00000111104.2
|
Psen2
|
presenilin 2 |
| chr4_+_84802592 | 2.07 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr13_+_4283729 | 2.06 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr16_+_90017634 | 2.05 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr7_+_81220987 | 2.03 |
ENSMUST00000165460.2
|
Whamm
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr2_+_30156733 | 2.00 |
ENSMUST00000113645.8
ENSMUST00000133877.8 ENSMUST00000139719.8 ENSMUST00000113643.8 ENSMUST00000150695.8 |
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr16_-_56984137 | 1.93 |
ENSMUST00000231733.2
|
Nit2
|
nitrilase family, member 2 |
| chr12_-_16660960 | 1.92 |
ENSMUST00000239165.2
ENSMUST00000111067.10 |
Lpin1
|
lipin 1 |
| chr15_+_76460550 | 1.91 |
ENSMUST00000162503.8
|
Adck5
|
aarF domain containing kinase 5 |
| chr17_+_34524841 | 1.90 |
ENSMUST00000235530.2
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr7_+_3620356 | 1.88 |
ENSMUST00000076657.11
ENSMUST00000108644.8 |
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr17_+_34524884 | 1.86 |
ENSMUST00000074557.11
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr9_+_109760856 | 1.85 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr2_+_30156523 | 1.84 |
ENSMUST00000091132.13
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr15_+_76460586 | 1.83 |
ENSMUST00000160784.8
|
Adck5
|
aarF domain containing kinase 5 |
| chr17_-_36353582 | 1.78 |
ENSMUST00000058801.15
ENSMUST00000080015.12 ENSMUST00000077960.7 |
H2-T22
|
histocompatibility 2, T region locus 22 |
| chr5_-_121523670 | 1.73 |
ENSMUST00000146185.2
ENSMUST00000042312.14 |
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr15_+_100768551 | 1.72 |
ENSMUST00000082209.13
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr11_-_116197994 | 1.72 |
ENSMUST00000124281.2
|
Exoc7
|
exocyst complex component 7 |
| chr7_-_140462187 | 1.67 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr7_-_119801327 | 1.63 |
ENSMUST00000033198.6
|
Crym
|
crystallin, mu |
| chr13_+_67080864 | 1.58 |
ENSMUST00000021990.4
|
Ptdss1
|
phosphatidylserine synthase 1 |
| chr4_-_149184259 | 1.56 |
ENSMUST00000103217.11
|
Pex14
|
peroxisomal biogenesis factor 14 |
| chr5_+_53424471 | 1.53 |
ENSMUST00000147148.5
|
Smim20
|
small integral membrane protein 20 |
| chr15_+_31224616 | 1.52 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr7_+_29883611 | 1.52 |
ENSMUST00000208441.2
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr14_-_34032311 | 1.50 |
ENSMUST00000111917.3
ENSMUST00000228704.2 |
Shld2
|
shieldin complex subunit 2 |
| chr6_+_94477294 | 1.50 |
ENSMUST00000061118.11
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr14_-_73613385 | 1.48 |
ENSMUST00000227454.2
|
Itm2b
|
integral membrane protein 2B |
| chr7_-_140462221 | 1.48 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr8_-_25506916 | 1.47 |
ENSMUST00000084035.12
ENSMUST00000208247.3 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr11_-_83469446 | 1.40 |
ENSMUST00000019266.6
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr11_+_119246376 | 1.40 |
ENSMUST00000050880.8
|
Slc26a11
|
solute carrier family 26, member 11 |
| chr16_-_17348882 | 1.39 |
ENSMUST00000231548.2
ENSMUST00000232041.2 ENSMUST00000231288.2 |
Thap7
|
THAP domain containing 7 |
| chr4_+_40722461 | 1.35 |
ENSMUST00000030118.10
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr11_-_53313950 | 1.34 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr17_-_28299569 | 1.34 |
ENSMUST00000129046.9
ENSMUST00000043925.16 |
Tcp11
|
t-complex protein 11 |
| chr4_+_116542741 | 1.32 |
ENSMUST00000135573.8
ENSMUST00000151129.8 |
Prdx1
|
peroxiredoxin 1 |
| chr6_-_124865155 | 1.32 |
ENSMUST00000024044.7
|
Cd4
|
CD4 antigen |
| chr8_-_84738761 | 1.31 |
ENSMUST00000191523.2
ENSMUST00000190457.2 ENSMUST00000185457.2 |
Misp3
|
MISP family member 3 |
| chr7_+_44240310 | 1.30 |
ENSMUST00000107906.6
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr17_+_31739089 | 1.29 |
ENSMUST00000064798.16
ENSMUST00000046288.16 ENSMUST00000191598.3 |
Ndufv3
|
NADH:ubiquinone oxidoreductase core subunit V3 |
| chr4_+_116543045 | 1.29 |
ENSMUST00000129315.8
ENSMUST00000106470.8 |
Prdx1
|
peroxiredoxin 1 |
| chr1_-_120001752 | 1.26 |
ENSMUST00000056089.8
|
Tmem37
|
transmembrane protein 37 |
| chr9_+_44309727 | 1.26 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr2_+_144398149 | 1.25 |
ENSMUST00000143573.8
ENSMUST00000028916.15 ENSMUST00000155258.2 |
Sec23b
|
SEC23 homolog B, COPII coat complex component |
| chr17_+_34138699 | 1.24 |
ENSMUST00000234320.2
|
Tapbp
|
TAP binding protein |
| chr1_+_46464625 | 1.23 |
ENSMUST00000189749.7
|
Dnah7c
|
dynein, axonemal, heavy chain 7C |
| chr19_+_44226155 | 1.21 |
ENSMUST00000237324.2
|
Scd3
|
stearoyl-coenzyme A desaturase 3 |
| chr12_-_16696958 | 1.19 |
ENSMUST00000238839.2
|
Lpin1
|
lipin 1 |
| chr1_+_167177545 | 1.18 |
ENSMUST00000028004.11
|
Aldh9a1
|
aldehyde dehydrogenase 9, subfamily A1 |
| chr7_-_27010068 | 1.17 |
ENSMUST00000125455.2
|
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chr16_+_20367327 | 1.16 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr9_-_72946980 | 1.15 |
ENSMUST00000184035.8
ENSMUST00000098566.5 |
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr1_-_40829801 | 1.14 |
ENSMUST00000039672.6
|
Mfsd9
|
major facilitator superfamily domain containing 9 |
| chr7_-_138511221 | 1.14 |
ENSMUST00000130500.8
ENSMUST00000106112.2 |
Bnip3
|
BCL2/adenovirus E1B interacting protein 3 |
| chr12_-_81531847 | 1.13 |
ENSMUST00000166723.8
ENSMUST00000110340.9 ENSMUST00000168463.8 ENSMUST00000169124.2 ENSMUST00000002757.11 |
Cox16
|
cytochrome c oxidase assembly protein 16 |
| chr11_-_48762170 | 1.11 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr8_-_25506756 | 1.11 |
ENSMUST00000084032.6
ENSMUST00000207132.2 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr2_+_144398226 | 1.11 |
ENSMUST00000155876.8
ENSMUST00000149697.3 |
Sec23b
|
SEC23 homolog B, COPII coat complex component |
| chr9_+_121211820 | 1.11 |
ENSMUST00000209995.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr13_-_54759145 | 1.09 |
ENSMUST00000091609.11
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr17_-_12726591 | 1.09 |
ENSMUST00000024595.4
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr11_+_83599841 | 1.09 |
ENSMUST00000001009.14
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr15_+_100768507 | 1.08 |
ENSMUST00000201518.4
ENSMUST00000200933.4 |
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr12_-_83609217 | 1.05 |
ENSMUST00000222448.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr16_+_48662894 | 1.03 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr1_+_157286124 | 1.02 |
ENSMUST00000193791.6
ENSMUST00000046743.11 ENSMUST00000119891.7 |
Cryzl2
|
crystallin zeta like 2 |
| chr19_-_20931566 | 1.00 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr18_-_13013030 | 1.00 |
ENSMUST00000119512.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr4_-_63779562 | 0.97 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr5_-_139805661 | 0.97 |
ENSMUST00000147328.2
|
Tmem184a
|
transmembrane protein 184a |
| chr4_+_98284128 | 0.96 |
ENSMUST00000107030.9
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr5_-_46014809 | 0.94 |
ENSMUST00000190036.7
ENSMUST00000189859.7 ENSMUST00000186633.3 ENSMUST00000016026.14 ENSMUST00000045586.13 ENSMUST00000238522.2 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr5_+_129924619 | 0.94 |
ENSMUST00000077320.3
|
Zbed5
|
zinc finger, BED type containing 5 |
| chr8_+_70243813 | 0.93 |
ENSMUST00000034326.7
|
Atp13a1
|
ATPase type 13A1 |
| chr5_-_113229445 | 0.93 |
ENSMUST00000131708.2
ENSMUST00000117143.8 ENSMUST00000119627.8 |
Crybb3
|
crystallin, beta B3 |
| chr7_+_46510627 | 0.90 |
ENSMUST00000014545.11
|
Ldhc
|
lactate dehydrogenase C |
| chr5_+_129924564 | 0.87 |
ENSMUST00000041466.14
|
Zbed5
|
zinc finger, BED type containing 5 |
| chr19_-_58849380 | 0.87 |
ENSMUST00000235263.2
|
Hspa12a
|
heat shock protein 12A |
| chr15_-_33687986 | 0.86 |
ENSMUST00000042021.5
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr4_+_43730034 | 0.85 |
ENSMUST00000131248.2
|
Spaar
|
small regulatory polypeptide of amino acid response |
| chr1_+_107350411 | 0.85 |
ENSMUST00000086690.6
|
Serpinb7
|
serine (or cysteine) peptidase inhibitor, clade B, member 7 |
| chr16_-_44153288 | 0.85 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr13_+_24118417 | 0.85 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr3_+_30800474 | 0.85 |
ENSMUST00000108262.10
|
Samd7
|
sterile alpha motif domain containing 7 |
| chr17_+_15230597 | 0.83 |
ENSMUST00000232446.2
|
Gm3435
|
predicted gene 3435 |
| chr2_-_180284468 | 0.82 |
ENSMUST00000037877.11
|
Tcfl5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr15_-_5093222 | 0.81 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr19_-_44534274 | 0.81 |
ENSMUST00000111985.2
ENSMUST00000063632.14 |
Sec31b
|
Sec31 homolog B (S. cerevisiae) |
| chr14_+_77274185 | 0.81 |
ENSMUST00000048208.10
ENSMUST00000095625.11 |
Ccdc122
|
coiled-coil domain containing 122 |
| chr5_-_92190859 | 0.80 |
ENSMUST00000069937.11
ENSMUST00000086978.12 |
Cdkl2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr13_-_24118139 | 0.79 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr14_-_77274056 | 0.79 |
ENSMUST00000062789.15
|
Lacc1
|
laccase domain containing 1 |
| chr18_+_69654992 | 0.78 |
ENSMUST00000201627.4
|
Tcf4
|
transcription factor 4 |
| chr11_+_70350436 | 0.78 |
ENSMUST00000039093.10
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr9_+_18320390 | 0.77 |
ENSMUST00000098973.3
|
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr7_+_140521450 | 0.77 |
ENSMUST00000164580.3
ENSMUST00000079403.11 |
Pgghg
|
protein glucosylgalactosylhydroxylysine glucosidase |
| chr17_+_15261896 | 0.76 |
ENSMUST00000226599.2
ENSMUST00000228518.2 ENSMUST00000226213.2 |
Ermard
|
ER membrane associated RNA degradation |
| chr3_+_40905216 | 0.76 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr12_-_75224099 | 0.75 |
ENSMUST00000042299.4
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr19_+_16416664 | 0.75 |
ENSMUST00000237350.2
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
| chr13_+_23214588 | 0.74 |
ENSMUST00000227652.2
ENSMUST00000227236.2 |
Vmn1r214
|
vomeronasal 1 receptor 214 |
| chr6_+_124973752 | 0.74 |
ENSMUST00000162000.4
|
Pianp
|
PILR alpha associated neural protein |
| chr14_-_20502285 | 0.73 |
ENSMUST00000056073.14
ENSMUST00000022349.14 ENSMUST00000022348.15 |
Cfap70
|
cilia and flagella associated protein 70 |
| chr1_+_86354045 | 0.72 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr2_-_66086919 | 0.72 |
ENSMUST00000125446.3
ENSMUST00000102718.10 |
Ttc21b
|
tetratricopeptide repeat domain 21B |
| chr2_-_127089540 | 0.72 |
ENSMUST00000174030.8
ENSMUST00000174863.8 |
Ciao1
|
cytosolic iron-sulfur protein assembly 1 |
| chr18_+_80299464 | 0.72 |
ENSMUST00000129043.8
ENSMUST00000151677.9 |
Slc66a2
|
solute carrier family 66 member 2 |
| chr17_+_34134873 | 0.71 |
ENSMUST00000172619.8
ENSMUST00000174463.2 |
Tapbp
Zbtb22
|
TAP binding protein zinc finger and BTB domain containing 22 |
| chr14_-_59632830 | 0.71 |
ENSMUST00000166912.3
|
Phf11c
|
PHD finger protein 11C |
| chr7_+_46510831 | 0.71 |
ENSMUST00000126004.3
|
Ldhc
|
lactate dehydrogenase C |
| chr8_+_46463633 | 0.70 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr13_+_19369097 | 0.69 |
ENSMUST00000103554.5
|
Trgv4
|
T cell receptor gamma, variable 4 |
| chr11_-_102297590 | 0.69 |
ENSMUST00000155104.8
ENSMUST00000130436.8 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr17_+_34866090 | 0.69 |
ENSMUST00000015605.15
|
Atf6b
|
activating transcription factor 6 beta |
| chr1_-_86353932 | 0.67 |
ENSMUST00000212541.2
|
Nmur1
|
neuromedin U receptor 1 |
| chr4_-_45012287 | 0.67 |
ENSMUST00000055028.9
ENSMUST00000180217.2 ENSMUST00000107817.3 |
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr6_+_124973644 | 0.66 |
ENSMUST00000032479.11
|
Pianp
|
PILR alpha associated neural protein |
| chr5_+_121987050 | 0.66 |
ENSMUST00000056654.8
ENSMUST00000198155.2 |
Pheta1
|
PH domain containing endocytic trafficking adaptor 1 |
| chr18_+_69654900 | 0.66 |
ENSMUST00000202057.4
|
Tcf4
|
transcription factor 4 |
| chr12_-_55045887 | 0.65 |
ENSMUST00000173529.2
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr11_+_58221569 | 0.65 |
ENSMUST00000073128.7
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr9_+_109760931 | 0.64 |
ENSMUST00000165876.8
|
Map4
|
microtubule-associated protein 4 |
| chr1_-_5140504 | 0.64 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr3_-_88362606 | 0.62 |
ENSMUST00000125526.8
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr17_+_8463886 | 0.62 |
ENSMUST00000231545.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr1_+_64729950 | 0.61 |
ENSMUST00000187170.7
|
Ccnyl1
|
cyclin Y-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 2.0 | 6.1 | GO:1904435 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 1.6 | 9.5 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.5 | 26.8 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 1.3 | 5.3 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 1.3 | 3.8 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 1.1 | 12.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 1.1 | 4.3 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 1.0 | 7.1 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.9 | 2.8 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.9 | 3.7 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.9 | 3.6 | GO:0015755 | fructose transport(GO:0015755) |
| 0.8 | 2.5 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.8 | 2.5 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.7 | 6.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.7 | 10.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 2.6 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.6 | 3.0 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.6 | 1.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.5 | 2.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.5 | 1.6 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.5 | 2.0 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.5 | 2.8 | GO:0050917 | sensory perception of umami taste(GO:0050917) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.4 | 1.3 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.4 | 3.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.4 | 3.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.4 | 3.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 2.0 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.4 | 3.4 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.4 | 11.4 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.4 | 1.5 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 0.8 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.3 | 2.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 3.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 6.0 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.2 | 2.9 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 3.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 3.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 1.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 1.3 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.2 | 1.9 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 0.6 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.2 | 1.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 2.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 2.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 4.4 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 3.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 1.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 1.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.2 | 0.5 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.2 | 1.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 3.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.4 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.1 | 1.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.4 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 2.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.7 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 3.3 | GO:0035455 | response to interferon-alpha(GO:0035455) negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 2.1 | GO:0043589 | Notch receptor processing(GO:0007220) skin morphogenesis(GO:0043589) |
| 0.1 | 1.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 2.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.2 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.1 | 2.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.4 | GO:0071895 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.1 | 7.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 1.7 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.5 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.4 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.1 | 0.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 2.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.4 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 0.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 12.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 2.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 5.6 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.2 | GO:1903465 | orbitofrontal cortex development(GO:0021769) motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.1 | 8.5 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.2 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 5.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.1 | GO:0015697 | organic cation transport(GO:0015695) quaternary ammonium group transport(GO:0015697) |
| 0.1 | 1.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 2.4 | GO:0015893 | drug transport(GO:0015893) |
| 0.1 | 0.9 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 1.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.7 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 2.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.3 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 1.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 0.1 | 0.2 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 1.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:2001076 | metanephric comma-shaped body morphogenesis(GO:0072278) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.0 | 0.4 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0098712 | L-glutamate import across plasma membrane(GO:0098712) |
| 0.0 | 1.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 1.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 2.8 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.2 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.5 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 0.5 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.5 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.6 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.6 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 1.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 5.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 2.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.0 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 1.3 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.3 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 2.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 3.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:1904306 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.8 | GO:0001832 | blastocyst growth(GO:0001832) |
| 0.0 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 4.1 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.2 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.4 | GO:0010524 | positive regulation of calcium ion transport into cytosol(GO:0010524) |
| 0.0 | 0.3 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 26.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.2 | 5.8 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.7 | 6.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 1.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.4 | 1.6 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.4 | 2.6 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.3 | 3.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 2.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 2.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 3.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 8.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.8 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 1.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 0.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 2.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.1 | 2.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 6.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 2.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.4 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 13.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 3.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.2 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.1 | 10.6 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 1.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 1.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 3.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 12.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 51.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 15.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 3.0 | 12.1 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 1.9 | 5.8 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 1.2 | 6.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.2 | 3.6 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 1.1 | 4.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.0 | 39.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.9 | 2.8 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.8 | 2.5 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.8 | 3.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.6 | 10.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 2.6 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.5 | 6.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 1.3 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.3 | 2.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 1.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 1.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 0.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 2.0 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.3 | 1.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.3 | 6.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 5.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 2.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 0.7 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.2 | 1.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 3.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 4.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 1.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 2.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 3.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 2.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 3.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 2.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.4 | GO:0001761 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.1 | 2.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 5.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 1.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 5.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 3.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 2.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 5.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 4.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 1.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 3.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.3 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.1 | 1.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 2.4 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 6.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.3 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 1.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 1.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 3.1 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 8.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 6.1 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.8 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 4.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 19.0 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 1.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 9.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.0 | 13.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.8 | 12.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 6.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 1.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.3 | 3.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.2 | 3.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 3.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 4.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 4.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 5.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.8 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |