Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
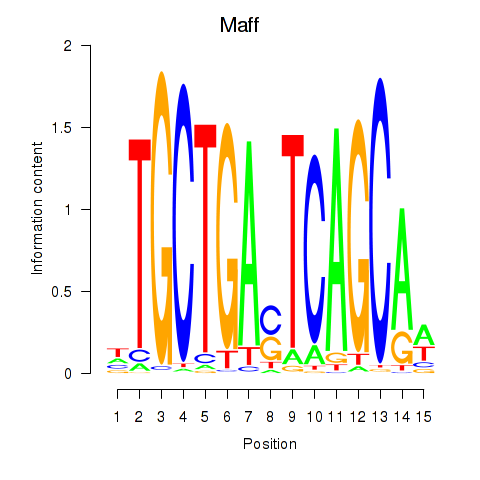
Results for Maff
Z-value: 2.60
Transcription factors associated with Maff
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maff
|
ENSMUSG00000042622.15 | Maff |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Maff | mm39_v1_chr15_+_79232137_79232264 | 0.82 | 1.2e-09 | Click! |
Activity profile of Maff motif
Sorted Z-values of Maff motif
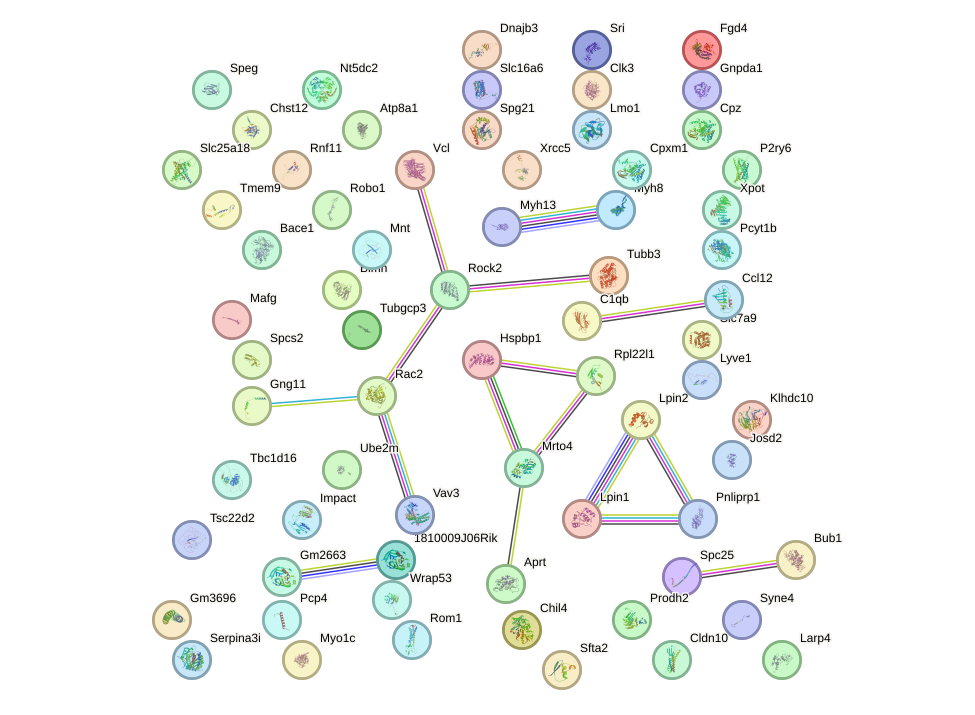
Network of associatons between targets according to the STRING database.
First level regulatory network of Maff
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_106126794 | 17.06 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr19_+_58717319 | 13.82 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr15_-_78456898 | 13.71 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr1_-_88133472 | 11.83 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr7_-_110462446 | 11.13 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr13_-_55677109 | 9.72 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr5_+_140491305 | 9.65 |
ENSMUST00000043050.9
ENSMUST00000124142.2 |
Chst12
|
carbohydrate sulfotransferase 12 |
| chr2_-_69036489 | 7.66 |
ENSMUST00000127243.8
ENSMUST00000149643.2 ENSMUST00000167875.9 ENSMUST00000005365.15 |
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr14_+_30853010 | 7.60 |
ENSMUST00000227096.2
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr5_-_67973195 | 6.84 |
ENSMUST00000141443.2
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr7_+_30193047 | 6.77 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr5_+_107112186 | 6.65 |
ENSMUST00000117196.9
ENSMUST00000031221.12 ENSMUST00000076467.13 |
Cdc7
|
cell division cycle 7 (S. cerevisiae) |
| chr11_-_109364424 | 6.21 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr2_-_69036472 | 6.18 |
ENSMUST00000112320.8
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chrX_+_92718695 | 5.91 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr1_+_135945705 | 5.72 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr2_-_127673738 | 5.71 |
ENSMUST00000028858.8
|
Bub1
|
BUB1, mitotic checkpoint serine/threonine kinase |
| chr11_+_67167950 | 5.51 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr7_-_108769719 | 5.14 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr6_-_40976413 | 5.09 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr7_+_44117444 | 5.09 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr7_+_44117511 | 4.99 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr1_+_135945798 | 4.71 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr5_+_8106527 | 4.64 |
ENSMUST00000148633.4
|
Sri
|
sorcin |
| chr5_+_103902426 | 4.52 |
ENSMUST00000153165.8
ENSMUST00000031256.6 |
Aff1
|
AF4/FMR2 family, member 1 |
| chr6_+_40941688 | 4.50 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr7_-_100613579 | 4.29 |
ENSMUST00000060174.6
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr8_+_72050292 | 4.07 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr8_-_12722099 | 3.91 |
ENSMUST00000000776.15
|
Tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr6_+_4003904 | 3.85 |
ENSMUST00000031670.10
|
Gng11
|
guanine nucleotide binding protein (G protein), gamma 11 |
| chr7_+_44117475 | 3.74 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr3_+_28859585 | 3.71 |
ENSMUST00000043867.11
ENSMUST00000194649.2 |
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr10_-_88440996 | 3.61 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr3_+_127584251 | 3.60 |
ENSMUST00000164447.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr10_-_88440869 | 3.56 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr7_+_30014235 | 3.55 |
ENSMUST00000054594.15
ENSMUST00000177078.8 ENSMUST00000176504.8 ENSMUST00000176304.8 |
Syne4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr7_+_44117404 | 3.48 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr10_+_127927443 | 3.13 |
ENSMUST00000238829.2
ENSMUST00000217851.2 ENSMUST00000220049.2 |
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr3_+_127584449 | 3.12 |
ENSMUST00000171621.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr11_+_75542902 | 2.88 |
ENSMUST00000102504.10
|
Myo1c
|
myosin IC |
| chr9_-_57673128 | 2.79 |
ENSMUST00000065330.8
|
Clk3
|
CDC-like kinase 3 |
| chr19_+_53186430 | 2.75 |
ENSMUST00000237099.2
|
Add3
|
adducin 3 (gamma) |
| chr8_-_123302187 | 2.59 |
ENSMUST00000213062.2
|
Aprt
|
adenine phosphoribosyl transferase |
| chr17_+_28059129 | 2.53 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr9_+_65368207 | 2.40 |
ENSMUST00000034955.8
ENSMUST00000213957.2 |
Spg21
|
SPG21, maspardin |
| chr17_+_28059099 | 2.40 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr15_+_99870661 | 2.39 |
ENSMUST00000100206.4
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr4_-_139079842 | 2.26 |
ENSMUST00000102503.10
|
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr7_-_12771554 | 2.26 |
ENSMUST00000125964.8
|
Ube2m
|
ubiquitin-conjugating enzyme E2M |
| chr14_+_119025306 | 2.21 |
ENSMUST00000047761.13
ENSMUST00000071546.14 |
Cldn10
|
claudin 10 |
| chr11_+_76836545 | 2.19 |
ENSMUST00000125145.8
|
Blmh
|
bleomycin hydrolase |
| chr17_+_28059036 | 2.17 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr4_-_109333866 | 2.11 |
ENSMUST00000030284.10
|
Rnf11
|
ring finger protein 11 |
| chr4_-_136613498 | 2.01 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr11_+_75542328 | 1.99 |
ENSMUST00000069057.13
|
Myo1c
|
myosin IC |
| chr14_+_20979466 | 1.83 |
ENSMUST00000022369.9
|
Vcl
|
vinculin |
| chr4_-_139079609 | 1.76 |
ENSMUST00000030513.13
ENSMUST00000155257.8 |
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr1_+_75358758 | 1.73 |
ENSMUST00000148515.8
ENSMUST00000113590.8 |
Speg
|
SPEG complex locus |
| chr7_+_35148188 | 1.71 |
ENSMUST00000118383.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr11_-_120520954 | 1.64 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr18_-_38471962 | 1.41 |
ENSMUST00000139885.2
ENSMUST00000235590.2 ENSMUST00000237487.2 ENSMUST00000063814.15 |
Gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr17_+_35960600 | 1.36 |
ENSMUST00000171166.3
|
Sfta2
|
surfactant associated 2 |
| chr7_+_35148461 | 1.34 |
ENSMUST00000118969.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr11_-_119119287 | 1.29 |
ENSMUST00000207655.2
ENSMUST00000036113.4 |
Tbc1d16
|
TBC1 domain family, member 16 |
| chr3_+_58322119 | 1.22 |
ENSMUST00000099090.7
ENSMUST00000199164.2 |
Tsc22d2
|
TSC22 domain family, member 2 |
| chr7_+_35148579 | 1.11 |
ENSMUST00000032703.10
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr5_-_35683035 | 1.06 |
ENSMUST00000038676.7
|
Cpz
|
carboxypeptidase Z |
| chr1_+_72346572 | 1.05 |
ENSMUST00000027379.10
|
Xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr16_+_72460029 | 1.00 |
ENSMUST00000023600.8
|
Robo1
|
roundabout guidance receptor 1 |
| chr11_+_74721733 | 0.97 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr18_+_13107535 | 0.90 |
ENSMUST00000234035.2
ENSMUST00000235053.2 |
Impact
|
impact, RWD domain protein |
| chr12_-_16696958 | 0.89 |
ENSMUST00000238839.2
|
Lpin1
|
lipin 1 |
| chr15_+_99870787 | 0.88 |
ENSMUST00000231160.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr3_+_109481223 | 0.86 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr17_+_71511642 | 0.84 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr10_-_121462219 | 0.81 |
ENSMUST00000039810.8
ENSMUST00000218004.2 |
Xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr14_-_36857202 | 0.75 |
ENSMUST00000165649.4
ENSMUST00000224769.2 |
Ghitm
|
growth hormone inducible transmembrane protein |
| chr11_+_76836330 | 0.74 |
ENSMUST00000021197.10
|
Blmh
|
bleomycin hydrolase |
| chr12_+_16944896 | 0.73 |
ENSMUST00000020904.8
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2 |
| chr11_-_69471056 | 0.72 |
ENSMUST00000132548.2
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr2_-_130239434 | 0.71 |
ENSMUST00000028897.8
|
Cpxm1
|
carboxypeptidase X 1 (M14 family) |
| chr19_-_8906686 | 0.60 |
ENSMUST00000096242.5
|
Rom1
|
rod outer segment membrane protein 1 |
| chr14_-_36857083 | 0.59 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr7_-_4687916 | 0.55 |
ENSMUST00000206306.2
ENSMUST00000205952.2 ENSMUST00000079970.6 |
Hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr16_-_16377982 | 0.50 |
ENSMUST00000161861.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr10_+_101517348 | 0.48 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr8_-_84420633 | 0.47 |
ENSMUST00000144258.8
|
Pkn1
|
protein kinase N1 |
| chr6_+_30401864 | 0.46 |
ENSMUST00000068240.13
ENSMUST00000068259.10 ENSMUST00000132581.8 |
Klhdc10
|
kelch domain containing 10 |
| chr14_+_15369152 | 0.38 |
ENSMUST00000167923.8
|
Gm3696
|
predicted gene 3696 |
| chr18_-_39652468 | 0.35 |
ENSMUST00000237944.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr9_+_45749869 | 0.35 |
ENSMUST00000078111.11
ENSMUST00000034591.11 |
Bace1
|
beta-site APP cleaving enzyme 1 |
| chr19_-_11301919 | 0.34 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr6_+_83771953 | 0.29 |
ENSMUST00000037376.14
|
Nagk
|
N-acetylglucosamine kinase |
| chr6_+_120750510 | 0.29 |
ENSMUST00000112682.4
|
Slc25a18
|
solute carrier family 25 (mitochondrial carrier), member 18 |
| chr12_+_104229376 | 0.25 |
ENSMUST00000109958.3
|
Serpina3i
|
serine (or cysteine) peptidase inhibitor, clade A, member 3I |
| chr2_-_20948230 | 0.18 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr15_+_99870714 | 0.17 |
ENSMUST00000230956.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr11_+_81992662 | 0.12 |
ENSMUST00000000194.4
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr8_+_124138163 | 0.10 |
ENSMUST00000071134.4
ENSMUST00000212743.2 |
Tubb3
|
tubulin, beta 3 class III |
| chr16_+_96295011 | 0.02 |
ENSMUST00000233816.2
|
Pcp4
|
Purkinje cell protein 4 |
| chr14_-_44239726 | 0.00 |
ENSMUST00000096869.3
|
Ang6
|
angiogenin, ribonuclease A family, member 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 2.3 | 6.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.9 | 17.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.4 | 13.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 1.1 | 5.7 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.1 | 6.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 1.0 | 4.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.8 | 7.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.8 | 6.8 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.7 | 5.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.7 | 2.9 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.7 | 1.4 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.6 | 2.6 | GO:0046084 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.6 | 4.9 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.6 | 11.1 | GO:0006026 | aminoglycan catabolic process(GO:0006026) glycosaminoglycan catabolic process(GO:0006027) |
| 0.5 | 4.6 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.5 | 4.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.4 | 7.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 1.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 3.9 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 3.5 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 0.9 | GO:1901561 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.3 | 3.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.3 | 1.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.3 | 5.5 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.2 | 0.8 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.7 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 4.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 17.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 13.4 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 0.7 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 1.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 2.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 5.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 0.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 3.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 6.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.9 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 3.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 2.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 7.6 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 2.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.2 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 1.4 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 3.5 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 1.0 | GO:0007569 | cell aging(GO:0007569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 13.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.9 | 5.7 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.2 | 4.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.0 | 3.9 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 1.0 | 4.9 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 3.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 7.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.5 | 5.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 7.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 3.5 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.1 | 6.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 4.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 11.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 13.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 3.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 10.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 6.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 4.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 1.9 | 17.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.8 | 7.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 1.5 | 5.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.1 | 4.3 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 1.0 | 4.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.9 | 2.6 | GO:0003999 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.8 | 2.3 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.7 | 13.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.5 | 1.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 11.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 7.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 10.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.3 | 7.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 3.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 6.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 2.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 3.1 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 6.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 17.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 4.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 9.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 1.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 3.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 6.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 2.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 11.8 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 9.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 3.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 8.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 5.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 11.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 9.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 14.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 2.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 7.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 12.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 19.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 6.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 6.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 4.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.8 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 1.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |