Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Meis1
Z-value: 1.03
Transcription factors associated with Meis1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meis1
|
ENSMUSG00000020160.19 | Meis1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis1 | mm39_v1_chr11_-_18968955_18968973 | -0.10 | 5.6e-01 | Click! |
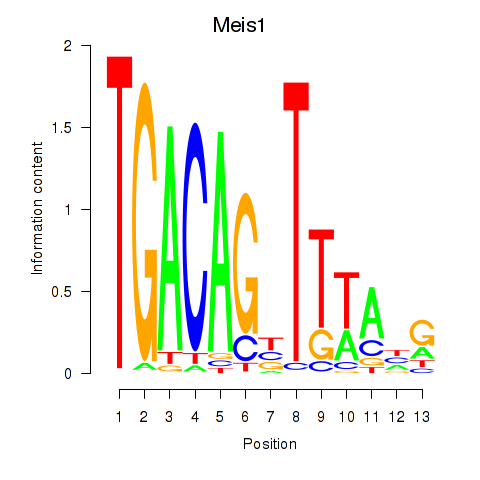
Activity profile of Meis1 motif
Sorted Z-values of Meis1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Meis1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_41098174 | 3.62 |
ENSMUST00000055327.8
|
Aqp3
|
aquaporin 3 |
| chr4_-_46404224 | 3.47 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr7_+_78563150 | 2.55 |
ENSMUST00000118867.8
|
Isg20
|
interferon-stimulated protein |
| chr7_+_78563184 | 2.50 |
ENSMUST00000121645.8
|
Isg20
|
interferon-stimulated protein |
| chr4_-_87724533 | 2.31 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr11_+_58808830 | 2.29 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr9_-_107863062 | 2.15 |
ENSMUST00000048568.6
|
Inka1
|
inka box actin regulator 1 |
| chr6_-_136918495 | 1.96 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr7_+_27147475 | 1.88 |
ENSMUST00000133750.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr11_-_69508706 | 1.85 |
ENSMUST00000005334.3
|
Shbg
|
sex hormone binding globulin |
| chr7_+_27147403 | 1.84 |
ENSMUST00000037399.16
ENSMUST00000108358.8 |
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr11_+_58808716 | 1.76 |
ENSMUST00000069941.13
|
Btnl10
|
butyrophilin-like 10 |
| chr5_+_44070486 | 1.59 |
ENSMUST00000122204.3
ENSMUST00000200338.2 |
Gm7879
|
predicted pseudogene 7879 |
| chr4_-_87724512 | 1.55 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr16_+_48692976 | 1.50 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr11_+_101207743 | 1.43 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr6_+_83003253 | 1.40 |
ENSMUST00000204891.2
|
M1ap
|
meiosis 1 associated protein |
| chr7_+_45084257 | 1.32 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr1_+_130659700 | 1.31 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr7_+_44221791 | 1.29 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase |
| chr11_+_117877118 | 1.26 |
ENSMUST00000132676.8
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr11_+_101333238 | 1.22 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chr18_+_36797113 | 1.20 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr15_-_78456898 | 1.18 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr9_+_86904067 | 1.17 |
ENSMUST00000168529.9
|
Cyb5r4
|
cytochrome b5 reductase 4 |
| chr11_+_101207021 | 1.15 |
ENSMUST00000142640.8
ENSMUST00000019470.14 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr15_+_61857390 | 1.09 |
ENSMUST00000159327.2
ENSMUST00000167731.8 |
Myc
|
myelocytomatosis oncogene |
| chr7_+_28140352 | 1.08 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr4_-_43578823 | 1.05 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr7_+_28140450 | 1.03 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr11_+_101333115 | 1.00 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr10_-_40018243 | 0.98 |
ENSMUST00000092566.8
ENSMUST00000213488.2 |
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
| chr1_+_156193607 | 0.92 |
ENSMUST00000102782.4
|
Gm2000
|
predicted gene 2000 |
| chrX_-_105884178 | 0.92 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chr19_-_37184692 | 0.91 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_+_76884182 | 0.88 |
ENSMUST00000041008.10
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr4_+_19818718 | 0.86 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr6_+_128376729 | 0.86 |
ENSMUST00000001561.12
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr7_-_115445352 | 0.85 |
ENSMUST00000206369.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr9_+_64188857 | 0.82 |
ENSMUST00000215031.2
ENSMUST00000213165.2 ENSMUST00000213289.2 ENSMUST00000216594.2 ENSMUST00000034964.7 |
Tipin
|
timeless interacting protein |
| chr6_-_133830613 | 0.81 |
ENSMUST00000203168.2
ENSMUST00000048032.5 |
Kap
|
kidney androgen regulated protein |
| chr8_-_71219299 | 0.80 |
ENSMUST00000222087.4
|
Ifi30
|
interferon gamma inducible protein 30 |
| chr11_-_109363406 | 0.78 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr17_+_6869070 | 0.78 |
ENSMUST00000092966.5
|
Dynlt1c
|
dynein light chain Tctex-type 1C |
| chr7_+_142025575 | 0.77 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr6_-_128868068 | 0.77 |
ENSMUST00000178918.2
ENSMUST00000160290.8 |
BC035044
|
cDNA sequence BC035044 |
| chr6_-_129853617 | 0.77 |
ENSMUST00000014687.11
ENSMUST00000122219.2 |
Klra17
|
killer cell lectin-like receptor, subfamily A, member 17 |
| chr7_+_142025817 | 0.76 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr6_-_30936013 | 0.73 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr8_-_79539838 | 0.73 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_-_115445315 | 0.71 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr6_+_128376844 | 0.71 |
ENSMUST00000120405.4
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr7_-_45084012 | 0.70 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr8_-_65489834 | 0.70 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr14_-_30741012 | 0.70 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr11_-_95733235 | 0.69 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr3_+_109030419 | 0.68 |
ENSMUST00000029477.11
|
Slc25a24
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 24 |
| chr2_-_168609110 | 0.68 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr12_-_69205882 | 0.66 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr6_+_113366207 | 0.66 |
ENSMUST00000204026.3
|
Ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr6_+_4747298 | 0.65 |
ENSMUST00000166678.2
ENSMUST00000176204.8 |
Peg10
|
paternally expressed 10 |
| chr5_+_114142842 | 0.64 |
ENSMUST00000161610.6
|
Dao
|
D-amino acid oxidase |
| chr17_-_71782296 | 0.63 |
ENSMUST00000127430.2
|
Smchd1
|
SMC hinge domain containing 1 |
| chr12_-_113542610 | 0.63 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr4_-_46566431 | 0.59 |
ENSMUST00000030021.14
ENSMUST00000107757.8 |
Coro2a
|
coronin, actin binding protein 2A |
| chr17_+_26895379 | 0.59 |
ENSMUST00000236299.2
ENSMUST00000167352.2 |
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr17_+_26895344 | 0.58 |
ENSMUST00000015719.16
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr1_-_75101826 | 0.58 |
ENSMUST00000152855.3
|
Nhej1
|
non-homologous end joining factor 1 |
| chr11_+_115455260 | 0.57 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr8_+_85583611 | 0.56 |
ENSMUST00000003906.13
ENSMUST00000109754.2 |
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chrX_+_154045439 | 0.55 |
ENSMUST00000026324.10
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr8_-_65489791 | 0.55 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr4_-_131988907 | 0.53 |
ENSMUST00000105964.2
|
Gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr4_-_43578636 | 0.53 |
ENSMUST00000130443.2
|
Gba2
|
glucosidase beta 2 |
| chr15_+_59246134 | 0.53 |
ENSMUST00000227173.2
ENSMUST00000079703.11 |
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr6_+_34389269 | 0.52 |
ENSMUST00000007449.9
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr7_-_19133783 | 0.52 |
ENSMUST00000047170.10
ENSMUST00000108459.9 |
Klc3
|
kinesin light chain 3 |
| chr4_+_154096188 | 0.51 |
ENSMUST00000169622.8
ENSMUST00000030894.15 |
Lrrc47
|
leucine rich repeat containing 47 |
| chr9_+_113760002 | 0.49 |
ENSMUST00000084885.12
ENSMUST00000009885.14 |
Ubp1
|
upstream binding protein 1 |
| chr3_+_157272504 | 0.49 |
ENSMUST00000041175.13
ENSMUST00000173533.2 |
Ptger3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr15_+_74586682 | 0.49 |
ENSMUST00000023265.5
|
Psca
|
prostate stem cell antigen |
| chr2_-_168608949 | 0.48 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr11_-_34674677 | 0.46 |
ENSMUST00000093193.12
ENSMUST00000101365.9 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr11_-_120538928 | 0.46 |
ENSMUST00000239158.2
ENSMUST00000026134.3 |
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr4_-_131988832 | 0.45 |
ENSMUST00000105965.8
|
Gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr7_-_33867940 | 0.45 |
ENSMUST00000102746.11
|
Uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr4_+_43578707 | 0.44 |
ENSMUST00000107886.9
ENSMUST00000117140.8 |
Rgp1
|
RAB6A GEF compex partner 1 |
| chr10_-_128329693 | 0.43 |
ENSMUST00000217776.2
ENSMUST00000219236.2 ENSMUST00000217733.2 ENSMUST00000218127.2 ENSMUST00000217969.2 ENSMUST00000164181.2 |
Myl6
|
myosin, light polypeptide 6, alkali, smooth muscle and non-muscle |
| chr1_+_43484895 | 0.43 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr9_+_77959206 | 0.43 |
ENSMUST00000024104.9
|
Gcm1
|
glial cells missing homolog 1 |
| chr2_-_28589675 | 0.43 |
ENSMUST00000124840.2
|
Spaca9
|
sperm acrosome associated 9 |
| chr11_-_70860778 | 0.43 |
ENSMUST00000108530.2
ENSMUST00000035283.11 ENSMUST00000108531.8 |
Nup88
|
nucleoporin 88 |
| chr11_-_58213939 | 0.43 |
ENSMUST00000108829.8
|
Zfp672
|
zinc finger protein 672 |
| chr6_+_56979285 | 0.42 |
ENSMUST00000079669.7
|
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr10_-_33662700 | 0.42 |
ENSMUST00000223295.2
|
Sult3a2
|
sulfotransferase family 3A, member 2 |
| chrX_-_72703330 | 0.42 |
ENSMUST00000114473.8
ENSMUST00000002087.14 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr1_+_118249558 | 0.42 |
ENSMUST00000027626.13
ENSMUST00000112688.10 |
Nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr9_+_75093177 | 0.40 |
ENSMUST00000129281.8
ENSMUST00000148144.8 ENSMUST00000130384.2 |
Myo5a
|
myosin VA |
| chr7_-_105230395 | 0.40 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr10_+_129153986 | 0.39 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr2_+_157298836 | 0.39 |
ENSMUST00000109529.2
|
Src
|
Rous sarcoma oncogene |
| chr15_+_34453432 | 0.39 |
ENSMUST00000060894.9
|
Erich5
|
glutamate rich 5 |
| chr8_+_34221861 | 0.38 |
ENSMUST00000170705.8
|
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr4_+_132262853 | 0.38 |
ENSMUST00000094657.10
ENSMUST00000105940.10 ENSMUST00000105939.10 ENSMUST00000150207.8 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr5_+_67765216 | 0.38 |
ENSMUST00000087241.7
|
Shisa3
|
shisa family member 3 |
| chr4_+_52607204 | 0.38 |
ENSMUST00000107671.2
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr5_+_143846782 | 0.37 |
ENSMUST00000148011.8
ENSMUST00000110709.7 |
Pms2
|
PMS1 homolog2, mismatch repair system component |
| chr6_+_4747356 | 0.37 |
ENSMUST00000176551.3
|
Peg10
|
paternally expressed 10 |
| chr8_+_34222058 | 0.36 |
ENSMUST00000167264.8
ENSMUST00000187392.7 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr4_-_154127119 | 0.35 |
ENSMUST00000047207.7
|
Ccdc27
|
coiled-coil domain containing 27 |
| chr10_+_99851679 | 0.35 |
ENSMUST00000130190.8
ENSMUST00000218200.2 ENSMUST00000020129.8 |
Kitl
|
kit ligand |
| chr3_-_106313406 | 0.35 |
ENSMUST00000029510.9
|
Chil6
|
chitinase-like 6 |
| chr12_-_80690573 | 0.34 |
ENSMUST00000166931.2
ENSMUST00000218364.2 |
Erh
|
ERH mRNA splicing and mitosis factor |
| chr17_-_79214328 | 0.34 |
ENSMUST00000042683.7
ENSMUST00000169544.8 |
Sult6b1
|
sulfotransferase family, cytosolic, 6B, member 1 |
| chr3_+_89366425 | 0.34 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr2_-_28589641 | 0.34 |
ENSMUST00000102877.8
|
Spaca9
|
sperm acrosome associated 9 |
| chr13_-_98628509 | 0.34 |
ENSMUST00000170205.2
|
Gm10320
|
predicted pseudogene 10320 |
| chr7_-_83384711 | 0.34 |
ENSMUST00000001792.12
|
Il16
|
interleukin 16 |
| chr17_+_25527616 | 0.34 |
ENSMUST00000015267.5
|
Prss28
|
protease, serine 28 |
| chr12_-_24731768 | 0.33 |
ENSMUST00000189849.7
ENSMUST00000186602.7 |
Cys1
|
cystin 1 |
| chr19_-_40371016 | 0.33 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr17_-_37523969 | 0.33 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr18_+_44237474 | 0.32 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr16_-_58910185 | 0.32 |
ENSMUST00000215647.2
|
Olfr191
|
olfactory receptor 191 |
| chr18_+_44237577 | 0.32 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr9_+_107173907 | 0.31 |
ENSMUST00000168260.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr2_+_85648823 | 0.31 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr11_+_78181667 | 0.31 |
ENSMUST00000046361.5
ENSMUST00000238934.2 |
Rskr
|
ribosomal protein S6 kinase related |
| chr2_-_180928867 | 0.31 |
ENSMUST00000130475.8
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr9_-_26717686 | 0.30 |
ENSMUST00000162702.8
ENSMUST00000040398.14 ENSMUST00000066560.13 |
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr11_-_94187893 | 0.30 |
ENSMUST00000132623.9
|
Luc7l3
|
LUC7-like 3 (S. cerevisiae) |
| chr17_-_37200425 | 0.30 |
ENSMUST00000172711.2
|
Trim40
|
tripartite motif-containing 40 |
| chr16_+_65317389 | 0.30 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr3_+_96537235 | 0.29 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr1_+_134333720 | 0.29 |
ENSMUST00000173908.8
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr15_-_36794741 | 0.29 |
ENSMUST00000110361.8
ENSMUST00000022894.14 ENSMUST00000110359.2 |
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr2_+_43638814 | 0.29 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chrX_-_162859429 | 0.29 |
ENSMUST00000134272.2
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B |
| chr6_+_24733239 | 0.29 |
ENSMUST00000031690.6
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chr5_+_124585244 | 0.29 |
ENSMUST00000198451.2
|
Kmt5a
|
lysine methyltransferase 5A |
| chr9_-_107483855 | 0.29 |
ENSMUST00000073448.12
ENSMUST00000194606.2 ENSMUST00000195662.6 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr2_+_93472657 | 0.28 |
ENSMUST00000042078.10
ENSMUST00000111254.2 |
Alx4
|
aristaless-like homeobox 4 |
| chr9_-_19968858 | 0.28 |
ENSMUST00000216538.2
ENSMUST00000212098.3 |
Olfr867
|
olfactory receptor 867 |
| chr16_-_18882012 | 0.28 |
ENSMUST00000199490.2
|
Iglj1
|
immunoglobulin lambda joining 1 |
| chr7_+_28563255 | 0.28 |
ENSMUST00000138272.8
|
Lgals7
|
lectin, galactose binding, soluble 7 |
| chr16_-_19132814 | 0.27 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chr16_-_26345493 | 0.27 |
ENSMUST00000165687.3
|
Tmem207
|
transmembrane protein 207 |
| chr9_-_119812042 | 0.27 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr7_-_92523396 | 0.27 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr8_+_106607507 | 0.27 |
ENSMUST00000040254.16
ENSMUST00000119261.8 |
Edc4
|
enhancer of mRNA decapping 4 |
| chr11_-_58692770 | 0.27 |
ENSMUST00000094156.12
ENSMUST00000239007.2 ENSMUST00000238886.2 ENSMUST00000060581.5 |
Fam183b
|
family with sequence similarity 183, member B |
| chr2_-_87467879 | 0.27 |
ENSMUST00000216082.2
|
Olfr1132
|
olfactory receptor 1132 |
| chr11_+_113575208 | 0.26 |
ENSMUST00000042227.15
ENSMUST00000123466.2 ENSMUST00000106621.4 |
D11Wsu47e
|
DNA segment, Chr 11, Wayne State University 47, expressed |
| chr11_+_70861007 | 0.26 |
ENSMUST00000018593.10
|
Rpain
|
RPA interacting protein |
| chr11_-_58214121 | 0.26 |
ENSMUST00000057836.12
ENSMUST00000064786.12 |
Zfp672
|
zinc finger protein 672 |
| chr12_-_113589576 | 0.25 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr3_-_87170903 | 0.25 |
ENSMUST00000090986.11
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr10_+_80084955 | 0.25 |
ENSMUST00000105364.8
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr5_-_136275407 | 0.24 |
ENSMUST00000196245.2
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr11_-_109718845 | 0.24 |
ENSMUST00000020941.11
|
1700012B07Rik
|
RIKEN cDNA 1700012B07 gene |
| chr7_+_127836502 | 0.24 |
ENSMUST00000044660.6
|
Armc5
|
armadillo repeat containing 5 |
| chr6_+_8520006 | 0.24 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr12_-_113561594 | 0.24 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr15_+_59246080 | 0.23 |
ENSMUST00000168722.3
|
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr17_-_45047521 | 0.23 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr4_+_101365144 | 0.22 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr2_-_86280934 | 0.22 |
ENSMUST00000216162.3
ENSMUST00000217586.3 ENSMUST00000213789.3 |
Olfr1065
|
olfactory receptor 1065 |
| chr3_+_116388600 | 0.22 |
ENSMUST00000198386.5
ENSMUST00000198311.5 ENSMUST00000197335.2 |
Sass6
|
SAS-6 centriolar assembly protein |
| chr10_+_101994841 | 0.22 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr8_-_95328348 | 0.22 |
ENSMUST00000212547.2
ENSMUST00000212507.2 ENSMUST00000034226.8 |
Psme3ip1
|
proteasome activator subunit 3 interacting protein 1 |
| chr17_+_21060706 | 0.22 |
ENSMUST00000232909.2
ENSMUST00000233670.2 ENSMUST00000233939.2 |
Vmn1r230
|
vomeronasal 1 receptor 230 |
| chr5_+_16139909 | 0.22 |
ENSMUST00000196750.2
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr9_-_44216892 | 0.22 |
ENSMUST00000034629.6
ENSMUST00000214660.2 ENSMUST00000216508.2 |
Hinfp
|
histone H4 transcription factor |
| chr19_+_13478481 | 0.21 |
ENSMUST00000214274.2
|
Olfr1477
|
olfactory receptor 1477 |
| chr12_-_113700190 | 0.20 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr5_+_67473069 | 0.20 |
ENSMUST00000202770.2
|
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr9_-_69668204 | 0.20 |
ENSMUST00000071281.5
|
Foxb1
|
forkhead box B1 |
| chr11_+_70861292 | 0.20 |
ENSMUST00000178822.8
ENSMUST00000108529.10 ENSMUST00000169965.8 ENSMUST00000167509.2 |
Rpain
|
RPA interacting protein |
| chr2_+_4980986 | 0.19 |
ENSMUST00000027978.7
ENSMUST00000195688.2 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr17_-_16051093 | 0.19 |
ENSMUST00000238157.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr12_-_108668520 | 0.19 |
ENSMUST00000167978.9
ENSMUST00000021691.6 |
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr12_-_113666198 | 0.19 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr2_+_180961507 | 0.18 |
ENSMUST00000098971.11
ENSMUST00000054622.15 ENSMUST00000108814.8 ENSMUST00000048608.16 ENSMUST00000108815.8 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr9_-_104214920 | 0.18 |
ENSMUST00000062723.14
ENSMUST00000215852.2 |
Acpp
|
acid phosphatase, prostate |
| chr6_-_67316645 | 0.18 |
ENSMUST00000117441.8
|
Il12rb2
|
interleukin 12 receptor, beta 2 |
| chr9_+_39818632 | 0.18 |
ENSMUST00000214904.2
|
Olfr229
|
olfactory receptor 229 |
| chr7_-_43885522 | 0.18 |
ENSMUST00000206686.2
ENSMUST00000037220.5 |
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr8_-_123338126 | 0.18 |
ENSMUST00000212319.2
|
Galns
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr2_+_4980923 | 0.18 |
ENSMUST00000167607.8
ENSMUST00000115010.9 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr12_+_91367764 | 0.18 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr2_-_113588983 | 0.17 |
ENSMUST00000099575.4
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr2_-_120561983 | 0.17 |
ENSMUST00000110701.8
ENSMUST00000110700.2 |
Cdan1
|
congenital dyserythropoietic anemia, type I (human) |
| chr1_-_169796709 | 0.17 |
ENSMUST00000027989.13
ENSMUST00000111353.4 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr14_+_60970355 | 0.17 |
ENSMUST00000162945.2
|
Spata13
|
spermatogenesis associated 13 |
| chr3_+_89366632 | 0.17 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr13_+_23953896 | 0.17 |
ENSMUST00000226039.2
|
Trim38
|
tripartite motif-containing 38 |
| chr12_-_50695881 | 0.17 |
ENSMUST00000002765.9
|
Prkd1
|
protein kinase D1 |
| chr13_+_38529062 | 0.16 |
ENSMUST00000171970.3
|
Bmp6
|
bone morphogenetic protein 6 |
| chr17_+_34617784 | 0.16 |
ENSMUST00000114175.8
ENSMUST00000139063.8 ENSMUST00000078615.12 |
BC051142
|
cDNA sequence BC051142 |
| chr2_+_26209755 | 0.16 |
ENSMUST00000066889.13
|
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.8 | 5.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.7 | 2.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.5 | 3.7 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 1.5 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 1.6 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.3 | 0.9 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.3 | 1.1 | GO:0046722 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 2.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 0.6 | GO:0036088 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 0.6 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.2 | 1.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 3.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.8 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 0.5 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of actin filament-based movement(GO:1903116) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.2 | 0.5 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 1.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.5 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.4 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.8 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.7 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 1.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 | 0.7 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 1.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 1.3 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.2 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 | 0.7 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.3 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.1 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.8 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.4 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.1 | 0.4 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 1.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.2 | GO:0042223 | positive regulation of type I hypersensitivity(GO:0001812) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.0 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.2 | GO:0061030 | axon target recognition(GO:0007412) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.7 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 1.2 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.4 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.8 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 1.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 3.7 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.1 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 2.2 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 3.0 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 1.0 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.7 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 0.6 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 0.6 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.4 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.1 | 0.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.8 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 1.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.4 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.1 | 0.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 5.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 3.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.4 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 1.0 | 5.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.7 | 3.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.5 | 1.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.4 | 1.3 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 2.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 2.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 1.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.5 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.2 | 0.7 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.2 | 0.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 2.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.6 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.3 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.1 | 1.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 0.7 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 2.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.8 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 2.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 2.0 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 3.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 3.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 5.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |