Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Meox1
Z-value: 1.05
Transcription factors associated with Meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox1
|
ENSMUSG00000001493.10 | Meox1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox1 | mm39_v1_chr11_-_101785181_101785200 | -0.32 | 6.0e-02 | Click! |
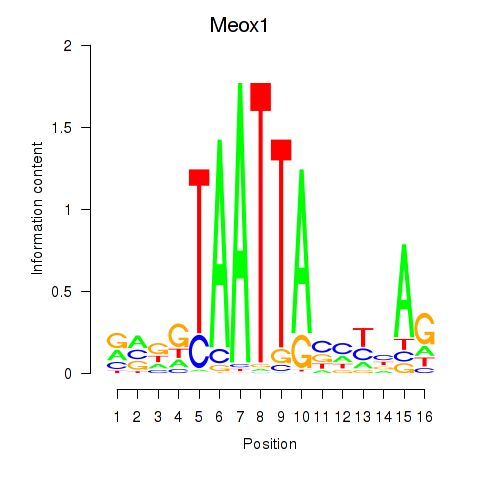
Activity profile of Meox1 motif
Sorted Z-values of Meox1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_107164347 | 7.22 |
ENSMUST00000082426.11
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr4_-_107164315 | 6.81 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr15_+_9335636 | 4.69 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr15_-_3612628 | 4.48 |
ENSMUST00000110698.9
|
Ghr
|
growth hormone receptor |
| chr10_+_127734384 | 4.20 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr15_-_3612078 | 3.19 |
ENSMUST00000161770.2
|
Ghr
|
growth hormone receptor |
| chr15_-_3612703 | 3.10 |
ENSMUST00000069451.11
|
Ghr
|
growth hormone receptor |
| chrM_+_10167 | 2.72 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr13_+_4283729 | 1.97 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr3_-_113371392 | 1.88 |
ENSMUST00000067980.12
|
Amy1
|
amylase 1, salivary |
| chr7_-_140590605 | 1.80 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr18_-_3281089 | 1.77 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr10_-_115197775 | 1.57 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr16_-_56984137 | 1.54 |
ENSMUST00000231733.2
|
Nit2
|
nitrilase family, member 2 |
| chrM_+_9459 | 1.49 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrM_+_9870 | 1.40 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr6_-_116084810 | 1.32 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr18_+_56565188 | 1.27 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr10_+_111342147 | 1.27 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1 |
| chr10_+_116111441 | 1.26 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr7_+_29931735 | 1.23 |
ENSMUST00000108193.2
ENSMUST00000108192.2 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chrM_+_7779 | 1.21 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr12_+_103498542 | 1.16 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr10_-_31321793 | 1.12 |
ENSMUST00000213639.2
ENSMUST00000215515.2 ENSMUST00000214644.2 ENSMUST00000213528.2 |
Tpd52l1
|
tumor protein D52-like 1 |
| chrM_+_8603 | 1.00 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr18_-_36877571 | 0.95 |
ENSMUST00000014438.5
|
Ndufa2
|
NADH:ubiquinone oxidoreductase subunit A2 |
| chrM_+_7758 | 0.94 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr7_+_101027390 | 0.94 |
ENSMUST00000084895.12
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr14_+_79086492 | 0.93 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr18_-_3280999 | 0.91 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr16_+_22676589 | 0.90 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr6_+_147377326 | 0.86 |
ENSMUST00000203659.3
ENSMUST00000032441.14 |
Ccdc91
|
coiled-coil domain containing 91 |
| chr5_-_23881353 | 0.85 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr2_+_69727563 | 0.83 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_-_70885877 | 0.82 |
ENSMUST00000090849.6
ENSMUST00000100037.9 ENSMUST00000112186.9 |
Mettl8
|
methyltransferase like 8 |
| chr9_+_123195986 | 0.81 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr5_-_87716882 | 0.77 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr6_-_125357756 | 0.77 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr7_+_29931309 | 0.76 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr7_-_12829100 | 0.75 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr13_-_53627110 | 0.75 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr2_+_179720416 | 0.73 |
ENSMUST00000087563.7
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr11_-_82781369 | 0.71 |
ENSMUST00000092844.13
ENSMUST00000021033.16 ENSMUST00000018985.15 |
Rad51d
|
RAD51 paralog D |
| chr13_+_34923589 | 0.71 |
ENSMUST00000221037.2
|
Fam50b
|
family with sequence similarity 50, member B |
| chr5_-_137015683 | 0.70 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr16_+_34815177 | 0.68 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chr3_-_54962899 | 0.68 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr9_+_27210500 | 0.68 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr3_-_54962922 | 0.68 |
ENSMUST00000197238.5
|
Ccna1
|
cyclin A1 |
| chr15_+_92495007 | 0.66 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr6_-_56546088 | 0.65 |
ENSMUST00000203372.3
|
Pde1c
|
phosphodiesterase 1C |
| chr7_-_119058489 | 0.65 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr11_+_100978103 | 0.65 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chrX_-_98514278 | 0.65 |
ENSMUST00000113797.4
ENSMUST00000113790.8 ENSMUST00000036354.7 ENSMUST00000167246.2 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr5_+_87814058 | 0.65 |
ENSMUST00000199506.5
ENSMUST00000197631.5 ENSMUST00000094641.9 |
Csn1s1
|
casein alpha s1 |
| chr17_-_45970238 | 0.58 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr18_-_38336893 | 0.57 |
ENSMUST00000194312.2
|
Pcdh1
|
protocadherin 1 |
| chr9_-_16412581 | 0.57 |
ENSMUST00000217308.3
|
Fat3
|
FAT atypical cadherin 3 |
| chr8_-_87307294 | 0.57 |
ENSMUST00000131423.8
ENSMUST00000152438.2 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr2_+_69727599 | 0.56 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr17_+_88748139 | 0.56 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr7_-_24423715 | 0.56 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr15_-_101801351 | 0.55 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr6_+_40619913 | 0.54 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr5_+_107551362 | 0.53 |
ENSMUST00000049146.12
|
Ephx4
|
epoxide hydrolase 4 |
| chr18_+_12874390 | 0.53 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr7_+_78922947 | 0.52 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr4_-_14621669 | 0.51 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr17_-_43003135 | 0.51 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr4_-_14621805 | 0.50 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr4_-_114991174 | 0.50 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr15_-_37458768 | 0.50 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr4_+_100336003 | 0.50 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr9_+_94551929 | 0.49 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr8_+_22682816 | 0.48 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr4_+_131649001 | 0.47 |
ENSMUST00000094666.4
|
Tmem200b
|
transmembrane protein 200B |
| chr9_+_40092216 | 0.46 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr11_+_116734104 | 0.46 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr7_+_6255709 | 0.45 |
ENSMUST00000121024.8
|
Epp13
|
epididymal protein 13 |
| chr11_+_84020475 | 0.44 |
ENSMUST00000133811.3
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr16_-_22676264 | 0.44 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr3_+_89153258 | 0.43 |
ENSMUST00000040888.12
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr11_+_43046476 | 0.43 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr14_-_7766684 | 0.42 |
ENSMUST00000225630.3
ENSMUST00000225979.3 ENSMUST00000226079.3 ENSMUST00000223607.3 ENSMUST00000223761.3 ENSMUST00000223981.3 ENSMUST00000224222.3 ENSMUST00000225175.3 ENSMUST00000225238.3 ENSMUST00000225232.3 |
Slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr10_-_85847697 | 0.41 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr11_-_99412084 | 0.41 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr2_-_164455520 | 0.41 |
ENSMUST00000094351.11
ENSMUST00000109338.2 |
Wfdc8
|
WAP four-disulfide core domain 8 |
| chr11_+_58062467 | 0.40 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr1_-_4479445 | 0.40 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr3_+_138019040 | 0.40 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr6_+_8948608 | 0.40 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr18_+_37433852 | 0.39 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr2_+_177760959 | 0.39 |
ENSMUST00000108916.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr17_-_50600620 | 0.39 |
ENSMUST00000010736.9
|
Dazl
|
deleted in azoospermia-like |
| chr16_+_21644692 | 0.39 |
ENSMUST00000232240.2
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr12_+_31488208 | 0.38 |
ENSMUST00000001254.6
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr16_-_48592319 | 0.37 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr15_-_8739893 | 0.36 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr18_+_12874368 | 0.36 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr19_-_11209797 | 0.36 |
ENSMUST00000186228.3
|
Ms4a12
|
membrane-spanning 4-domains, subfamily A, member 12 |
| chrX_+_36675081 | 0.36 |
ENSMUST00000115179.4
|
Rhox2d
|
reproductive homeobox 2D |
| chr7_+_126549692 | 0.36 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr10_+_115854118 | 0.36 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr11_+_73262072 | 0.36 |
ENSMUST00000078952.9
ENSMUST00000120401.9 ENSMUST00000170592.4 |
Olfr376
|
olfactory receptor 376 |
| chr13_-_19521337 | 0.35 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chrX_+_36712105 | 0.35 |
ENSMUST00000072167.10
ENSMUST00000184746.2 |
Rhox2e
|
reproductive homeobox 2E |
| chr6_-_41752111 | 0.34 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr14_-_48900192 | 0.34 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr14_-_109151590 | 0.34 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr10_-_81186025 | 0.33 |
ENSMUST00000122993.8
|
Hmg20b
|
high mobility group 20B |
| chr5_-_96312006 | 0.33 |
ENSMUST00000137207.2
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr10_-_81186137 | 0.32 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr8_+_65399831 | 0.32 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr2_+_164455438 | 0.32 |
ENSMUST00000094346.3
|
Wfdc6b
|
WAP four-disulfide core domain 6B |
| chr12_-_99529767 | 0.31 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr7_-_29931612 | 0.30 |
ENSMUST00000006254.6
|
Tbcb
|
tubulin folding cofactor B |
| chr4_-_14621497 | 0.30 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr4_-_118549953 | 0.30 |
ENSMUST00000216226.2
|
Olfr1342
|
olfactory receptor 1342 |
| chr3_-_120965327 | 0.30 |
ENSMUST00000170781.2
ENSMUST00000039761.12 ENSMUST00000106467.8 ENSMUST00000106466.10 ENSMUST00000164925.9 |
Rwdd3
|
RWD domain containing 3 |
| chr2_+_89842475 | 0.29 |
ENSMUST00000214382.2
ENSMUST00000217065.3 |
Olfr1263
|
olfactory receptor 1263 |
| chr3_-_121056944 | 0.29 |
ENSMUST00000128909.8
ENSMUST00000029777.14 |
Tlcd4
|
TLC domain containing 4 |
| chr4_-_138351313 | 0.29 |
ENSMUST00000030533.12
|
Vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr4_-_118795809 | 0.29 |
ENSMUST00000215312.3
|
Olfr1328
|
olfactory receptor 1328 |
| chr1_-_163552693 | 0.29 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr14_+_54198389 | 0.28 |
ENSMUST00000103678.4
|
Trdv2-2
|
T cell receptor delta variable 2-2 |
| chr10_-_81186222 | 0.28 |
ENSMUST00000020454.11
ENSMUST00000105324.9 ENSMUST00000154609.3 ENSMUST00000105323.8 |
Hmg20b
|
high mobility group 20B |
| chr1_-_75208734 | 0.28 |
ENSMUST00000180101.3
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr9_+_19247753 | 0.28 |
ENSMUST00000215572.3
ENSMUST00000213344.2 |
Olfr845
|
olfactory receptor 845 |
| chr9_+_24194729 | 0.27 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr17_+_38032699 | 0.27 |
ENSMUST00000207771.3
|
Olfr120
|
olfactory receptor 120 |
| chrX_-_142716085 | 0.26 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr11_-_65160810 | 0.25 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chr11_+_88964667 | 0.25 |
ENSMUST00000100619.11
|
Gm525
|
predicted gene 525 |
| chr17_+_93506590 | 0.25 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr4_-_114991478 | 0.24 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr3_+_102927901 | 0.23 |
ENSMUST00000198180.5
ENSMUST00000197827.5 ENSMUST00000199240.5 ENSMUST00000199420.5 ENSMUST00000199571.5 ENSMUST00000197488.5 |
Csde1
|
cold shock domain containing E1, RNA binding |
| chr9_+_35580920 | 0.23 |
ENSMUST00000118254.2
|
Pate2
|
prostate and testis expressed 2 |
| chr1_+_192856044 | 0.23 |
ENSMUST00000193307.2
|
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
| chr8_-_55177510 | 0.23 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr13_+_22534534 | 0.22 |
ENSMUST00000226909.2
ENSMUST00000227167.2 ENSMUST00000226786.2 |
Vmn1r198
|
vomeronasal 1 receptor 198 |
| chr17_+_93506435 | 0.22 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr10_-_128885867 | 0.22 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr10_+_57527084 | 0.21 |
ENSMUST00000175852.2
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr12_-_104439589 | 0.21 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr4_+_102843540 | 0.20 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr7_-_103113358 | 0.20 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chr10_+_57527073 | 0.20 |
ENSMUST00000066028.13
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr8_+_84262409 | 0.20 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr13_+_22508759 | 0.19 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr3_-_15491482 | 0.19 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr9_+_35580941 | 0.19 |
ENSMUST00000217565.2
|
Pate2
|
prostate and testis expressed 2 |
| chr8_+_12981662 | 0.19 |
ENSMUST00000238472.2
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr11_+_115802828 | 0.18 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr18_+_3382968 | 0.18 |
ENSMUST00000025073.12
|
Cul2
|
cullin 2 |
| chr11_-_65160767 | 0.18 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chrX_-_142716200 | 0.17 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr11_+_60956624 | 0.17 |
ENSMUST00000041944.9
ENSMUST00000108717.3 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr1_-_158183894 | 0.17 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr5_-_99091681 | 0.17 |
ENSMUST00000162619.8
ENSMUST00000162147.6 |
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr6_-_123395075 | 0.17 |
ENSMUST00000172199.3
|
Vmn2r20
|
vomeronasal 2, receptor 20 |
| chr7_-_102638531 | 0.16 |
ENSMUST00000215606.2
|
Olfr578
|
olfactory receptor 578 |
| chr2_+_59442378 | 0.16 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr11_-_114851243 | 0.16 |
ENSMUST00000092466.13
ENSMUST00000061637.4 |
Cd300c
|
CD300C molecule |
| chr10_+_127919142 | 0.16 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr14_+_54082691 | 0.16 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr15_-_8740218 | 0.16 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr5_-_84565218 | 0.15 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr18_+_52748978 | 0.15 |
ENSMUST00000072666.4
ENSMUST00000209270.2 |
Zfp474
|
zinc finger protein 474 |
| chr5_+_138185747 | 0.15 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr19_-_50667079 | 0.15 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr6_-_124756645 | 0.15 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr11_+_73241609 | 0.14 |
ENSMUST00000120137.3
|
Olfr20
|
olfactory receptor 20 |
| chr4_+_8690398 | 0.14 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr14_+_26414422 | 0.14 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr15_+_98329106 | 0.14 |
ENSMUST00000215320.3
|
Olfr282
|
olfactory receptor 282 |
| chr6_-_89825033 | 0.14 |
ENSMUST00000226436.2
|
Vmn1r42
|
vomeronasal 1 receptor 42 |
| chr9_+_38491474 | 0.13 |
ENSMUST00000217160.3
|
Olfr912
|
olfactory receptor 912 |
| chr6_+_56933451 | 0.13 |
ENSMUST00000096612.4
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr11_+_49033101 | 0.13 |
ENSMUST00000203377.4
ENSMUST00000214195.3 |
Olfr1395
|
olfactory receptor 1395 |
| chr14_-_50521663 | 0.13 |
ENSMUST00000213701.2
|
Olfr732
|
olfactory receptor 732 |
| chr4_+_62443606 | 0.13 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr18_+_46874970 | 0.13 |
ENSMUST00000224622.2
|
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr3_+_14011445 | 0.13 |
ENSMUST00000192209.6
ENSMUST00000171075.8 ENSMUST00000108372.4 |
Ralyl
|
RALY RNA binding protein-like |
| chr11_-_59484115 | 0.13 |
ENSMUST00000215626.2
|
Olfr223
|
olfactory receptor 223 |
| chr3_-_58729732 | 0.12 |
ENSMUST00000191233.4
|
Mindy4b-ps
|
MINDY lysine 48 deubiquitinase 4B, pseudogene |
| chr9_-_70842090 | 0.12 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr10_+_81554753 | 0.12 |
ENSMUST00000085664.6
|
Zfp433
|
zinc finger protein 433 |
| chr1_+_66507523 | 0.12 |
ENSMUST00000061620.17
ENSMUST00000212557.3 |
Unc80
|
unc-80, NALCN activator |
| chr12_+_38830812 | 0.12 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr19_-_11838430 | 0.11 |
ENSMUST00000214796.2
|
Olfr1418
|
olfactory receptor 1418 |
| chrX_-_91643178 | 0.11 |
ENSMUST00000113955.2
|
Mageb18
|
MAGE family member B18 |
| chr13_-_23041731 | 0.11 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chrX_-_100129626 | 0.10 |
ENSMUST00000113710.8
|
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr8_-_41494890 | 0.10 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chrX_+_135723420 | 0.10 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr3_+_125474628 | 0.10 |
ENSMUST00000174648.6
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr15_+_25774070 | 0.10 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr10_+_41179966 | 0.10 |
ENSMUST00000173494.4
|
Ak9
|
adenylate kinase 9 |
| chr9_+_20193647 | 0.10 |
ENSMUST00000071725.4
ENSMUST00000212983.3 |
Olfr39
|
olfactory receptor 39 |
| chr13_+_22656093 | 0.09 |
ENSMUST00000226330.2
ENSMUST00000226965.2 |
Vmn1r201
|
vomeronasal 1 receptor 201 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.3 | 1.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 1.2 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.3 | 0.8 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.2 | 0.7 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.2 | 0.7 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.2 | 2.0 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.5 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:2000722 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 4.7 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.1 | 14.0 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 2.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.9 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.5 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.7 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.4 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.7 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 1.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.3 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 2.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 1.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.9 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 2.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 2.4 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.4 | GO:0046666 | retinal cell programmed cell death(GO:0046666) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.7 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 1.5 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.4 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.9 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 1.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.4 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 1.4 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.2 | 0.9 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.2 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 2.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 2.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.2 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 3.5 | 14.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 0.8 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.3 | 2.4 | GO:0016160 | amylase activity(GO:0016160) |
| 0.2 | 6.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 4.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 4.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.8 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.7 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 2.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.4 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 2.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 14.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 10.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 3.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |