Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Meox2
Z-value: 0.82
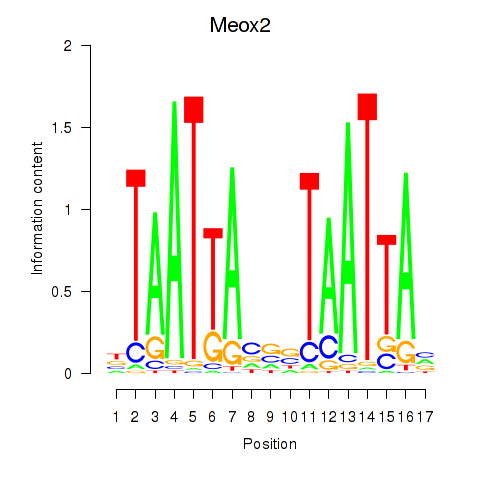
Transcription factors associated with Meox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox2
|
ENSMUSG00000036144.7 | Meox2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox2 | mm39_v1_chr12_+_37158532_37158545 | -0.27 | 1.1e-01 | Click! |
Activity profile of Meox2 motif
Sorted Z-values of Meox2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_93857899 | 4.35 |
ENSMUST00000034189.17
|
Ces1c
|
carboxylesterase 1C |
| chr6_-_41012435 | 3.89 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr8_-_93806593 | 3.51 |
ENSMUST00000109582.3
|
Ces1b
|
carboxylesterase 1B |
| chr6_+_41279199 | 2.49 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr13_+_4486105 | 2.22 |
ENSMUST00000156277.2
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chrM_+_10167 | 1.70 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr6_-_78355834 | 1.57 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr5_-_87240405 | 1.41 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr6_+_41331039 | 1.33 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr2_+_67935015 | 1.30 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr6_+_42222841 | 1.24 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr11_-_109986763 | 1.24 |
ENSMUST00000046223.14
ENSMUST00000106664.10 ENSMUST00000106662.2 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr1_-_162726234 | 1.21 |
ENSMUST00000111510.8
ENSMUST00000045902.13 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr13_-_4659120 | 1.20 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr11_-_109986804 | 1.19 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr3_+_20011405 | 1.18 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr9_-_51240201 | 1.11 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chrM_+_9870 | 1.09 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr4_-_115353326 | 1.08 |
ENSMUST00000030487.3
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chr9_-_21838758 | 0.94 |
ENSMUST00000046831.11
ENSMUST00000238930.2 |
Tmem205
|
transmembrane protein 205 |
| chr17_-_34506744 | 0.89 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr1_+_130754413 | 0.83 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr15_-_60793115 | 0.79 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr1_+_87983099 | 0.78 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr15_+_31225302 | 0.74 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr1_+_87983189 | 0.72 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr3_-_113368407 | 0.72 |
ENSMUST00000106540.8
|
Amy1
|
amylase 1, salivary |
| chr13_-_19491721 | 0.69 |
ENSMUST00000103561.3
|
Trgc2
|
T cell receptor gamma, constant 2 |
| chr18_+_60936910 | 0.68 |
ENSMUST00000097563.9
ENSMUST00000050487.16 ENSMUST00000167610.2 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr2_+_67578556 | 0.66 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chrM_+_7758 | 0.64 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr11_+_118324652 | 0.61 |
ENSMUST00000106286.3
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr11_-_43792013 | 0.60 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr15_+_31224555 | 0.60 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr2_+_148523118 | 0.60 |
ENSMUST00000131292.2
|
Gzf1
|
GDNF-inducible zinc finger protein 1 |
| chr5_+_90608751 | 0.58 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr17_+_45817750 | 0.57 |
ENSMUST00000024733.9
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr10_+_128106414 | 0.56 |
ENSMUST00000085708.3
ENSMUST00000105238.10 |
Stat2
|
signal transducer and activator of transcription 2 |
| chrM_+_3906 | 0.54 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr2_+_19376447 | 0.52 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr2_+_148522943 | 0.51 |
ENSMUST00000028928.8
|
Gzf1
|
GDNF-inducible zinc finger protein 1 |
| chrM_+_14138 | 0.50 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr8_+_71995554 | 0.50 |
ENSMUST00000034272.9
|
Mvb12a
|
multivesicular body subunit 12A |
| chrM_+_11735 | 0.47 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrM_-_14061 | 0.46 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr5_-_147831610 | 0.46 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr12_+_80690985 | 0.46 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr9_+_21838767 | 0.46 |
ENSMUST00000006403.7
ENSMUST00000170304.9 ENSMUST00000216710.2 |
Ccdc159
|
coiled-coil domain containing 159 |
| chrM_+_9459 | 0.45 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr5_-_125371162 | 0.45 |
ENSMUST00000127148.2
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr12_+_80691275 | 0.44 |
ENSMUST00000217889.2
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr14_+_73376192 | 0.44 |
ENSMUST00000171070.8
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr13_-_30039613 | 0.44 |
ENSMUST00000091674.12
ENSMUST00000006353.14 |
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr17_+_46807637 | 0.44 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr7_-_127897253 | 0.42 |
ENSMUST00000033044.16
|
Rusf1
|
RUS family member 1 |
| chr11_-_12414850 | 0.39 |
ENSMUST00000109650.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr17_-_15784582 | 0.38 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chr6_-_30693675 | 0.37 |
ENSMUST00000169422.8
ENSMUST00000115131.8 ENSMUST00000115130.3 ENSMUST00000031810.15 |
Cep41
|
centrosomal protein 41 |
| chr19_-_57170725 | 0.35 |
ENSMUST00000133369.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr16_-_19079594 | 0.33 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr14_-_55950545 | 0.33 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr9_+_94551929 | 0.33 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr8_+_22996233 | 0.33 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr4_+_102278715 | 0.31 |
ENSMUST00000106904.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr6_+_34575435 | 0.30 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chrM_+_7779 | 0.29 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr9_+_121245036 | 0.29 |
ENSMUST00000211187.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr5_-_108017019 | 0.28 |
ENSMUST00000128723.8
ENSMUST00000124034.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr3_+_108093645 | 0.28 |
ENSMUST00000050909.7
ENSMUST00000106659.3 ENSMUST00000106656.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr13_+_19398273 | 0.28 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr5_-_138617952 | 0.28 |
ENSMUST00000063262.11
ENSMUST00000085852.11 ENSMUST00000110905.9 |
Zfp68
|
zinc finger protein 68 |
| chr18_-_66155651 | 0.27 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr6_+_41511248 | 0.27 |
ENSMUST00000192366.2
ENSMUST00000103286.2 |
Trbj1-3
|
T cell receptor beta joining 1-3 |
| chr16_-_56688024 | 0.26 |
ENSMUST00000232373.2
|
Tmem45a
|
transmembrane protein 45a |
| chr6_+_94477294 | 0.26 |
ENSMUST00000061118.11
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr15_+_6451721 | 0.26 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr16_+_4457805 | 0.25 |
ENSMUST00000038770.4
|
Vasn
|
vasorin |
| chr8_-_37200051 | 0.25 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr13_+_46822992 | 0.25 |
ENSMUST00000099547.4
|
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr10_-_128640232 | 0.25 |
ENSMUST00000051011.14
|
Tmem198b
|
transmembrane protein 198b |
| chr11_+_119493806 | 0.25 |
ENSMUST00000026671.13
|
Rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr7_+_100435548 | 0.24 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A |
| chr8_-_41469786 | 0.22 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr1_-_4479445 | 0.22 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr2_+_136733421 | 0.22 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr9_+_108566513 | 0.22 |
ENSMUST00000192344.2
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr18_+_38429688 | 0.22 |
ENSMUST00000170811.8
ENSMUST00000072376.13 ENSMUST00000237903.2 ENSMUST00000236116.2 ENSMUST00000237824.2 ENSMUST00000236982.2 ENSMUST00000235549.2 ENSMUST00000237416.2 ENSMUST00000237089.2 |
Rnf14
|
ring finger protein 14 |
| chrX_+_41157242 | 0.21 |
ENSMUST00000115095.9
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr5_-_108943211 | 0.20 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr16_+_16688692 | 0.20 |
ENSMUST00000232547.2
|
Top3b
|
topoisomerase (DNA) III beta |
| chr16_+_42727926 | 0.20 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr5_+_138185747 | 0.20 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr11_+_49184124 | 0.19 |
ENSMUST00000189851.3
|
Olfr1392
|
olfactory receptor 1392 |
| chr2_-_136733282 | 0.19 |
ENSMUST00000028730.13
ENSMUST00000110089.9 ENSMUST00000227806.2 |
Mkks
ENSMUSG00000115423.3
|
McKusick-Kaufman syndrome novel protein |
| chr4_-_150087587 | 0.18 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr12_-_81531847 | 0.18 |
ENSMUST00000166723.8
ENSMUST00000110340.9 ENSMUST00000168463.8 ENSMUST00000169124.2 ENSMUST00000002757.11 |
Cox16
|
cytochrome c oxidase assembly protein 16 |
| chr8_+_57004125 | 0.17 |
ENSMUST00000110322.9
ENSMUST00000040218.13 ENSMUST00000210863.2 |
Fbxo8
|
F-box protein 8 |
| chr1_-_155910567 | 0.17 |
ENSMUST00000141878.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr17_-_6129968 | 0.16 |
ENSMUST00000024570.6
ENSMUST00000097432.10 |
Serac1
|
serine active site containing 1 |
| chr5_+_24598633 | 0.16 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr6_+_41510788 | 0.15 |
ENSMUST00000103284.2
|
Trbj1-1
|
T cell receptor beta joining 1-1 |
| chr5_-_99400698 | 0.15 |
ENSMUST00000031276.15
ENSMUST00000168092.8 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr8_+_45960855 | 0.14 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr10_-_127358231 | 0.14 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_67913949 | 0.14 |
ENSMUST00000032770.4
ENSMUST00000207874.2 |
Pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr7_+_6733561 | 0.14 |
ENSMUST00000200535.6
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr2_+_136734088 | 0.14 |
ENSMUST00000099311.9
|
Slx4ip
|
SLX4 interacting protein |
| chr10_-_43934774 | 0.14 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr17_-_50600620 | 0.14 |
ENSMUST00000010736.9
|
Dazl
|
deleted in azoospermia-like |
| chr2_-_45000250 | 0.13 |
ENSMUST00000201211.4
ENSMUST00000177302.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_165230165 | 0.13 |
ENSMUST00000103084.4
|
Zfp334
|
zinc finger protein 334 |
| chr11_-_70405429 | 0.13 |
ENSMUST00000021179.4
|
Vmo1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr15_+_80120036 | 0.13 |
ENSMUST00000226485.2
|
ENSMUSG00000115798.2
|
novel protein |
| chr1_+_180878797 | 0.13 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr5_+_104318542 | 0.13 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr7_+_6733684 | 0.13 |
ENSMUST00000197117.5
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr1_-_30988381 | 0.13 |
ENSMUST00000232841.2
|
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr1_-_20854490 | 0.13 |
ENSMUST00000039046.10
|
Il17f
|
interleukin 17F |
| chr11_+_58062467 | 0.13 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chrM_+_8603 | 0.12 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr3_-_96104886 | 0.12 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chr4_-_144447974 | 0.12 |
ENSMUST00000036876.8
|
AAdacl4fm3
|
AADACL4 family member 3 |
| chr17_-_40630096 | 0.12 |
ENSMUST00000026498.5
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr17_+_64203017 | 0.11 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr16_-_16962256 | 0.11 |
ENSMUST00000115711.10
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr2_-_112198366 | 0.11 |
ENSMUST00000028551.4
|
Emc4
|
ER membrane protein complex subunit 4 |
| chr14_-_118370144 | 0.10 |
ENSMUST00000022727.10
ENSMUST00000228543.2 |
Tgds
|
TDP-glucose 4,6-dehydratase |
| chr2_+_129642371 | 0.10 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr11_-_105828624 | 0.10 |
ENSMUST00000183493.8
|
Cyb561
|
cytochrome b-561 |
| chr4_+_126971167 | 0.10 |
ENSMUST00000046751.13
ENSMUST00000094713.4 |
Zmym6
|
zinc finger, MYM-type 6 |
| chr2_+_26471062 | 0.10 |
ENSMUST00000238951.2
ENSMUST00000166920.10 |
Egfl7
|
EGF-like domain 7 |
| chr17_+_47083561 | 0.10 |
ENSMUST00000071430.7
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chrX_-_88453295 | 0.10 |
ENSMUST00000113959.8
ENSMUST00000113960.3 |
Dcaf8l
|
DDB1 and CUL4 associated factor 8 like |
| chr14_-_36820304 | 0.10 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr7_-_139628991 | 0.10 |
ENSMUST00000172775.4
ENSMUST00000055353.9 |
Msx3
|
msh homeobox 3 |
| chr2_-_150934270 | 0.10 |
ENSMUST00000109890.2
|
Gm14151
|
predicted gene 14151 |
| chrX_-_95000496 | 0.10 |
ENSMUST00000079987.13
ENSMUST00000113864.3 |
Las1l
|
LAS1-like (S. cerevisiae) |
| chr5_+_107977965 | 0.09 |
ENSMUST00000072578.8
|
Ube2d2b
|
ubiquitin-conjugating enzyme E2D 2B |
| chr3_+_14598848 | 0.09 |
ENSMUST00000108370.9
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr1_+_163607143 | 0.09 |
ENSMUST00000077642.12
ENSMUST00000027877.7 |
Kifap3
|
kinesin-associated protein 3 |
| chr3_+_92223927 | 0.09 |
ENSMUST00000061038.4
|
Sprr2b
|
small proline-rich protein 2B |
| chr13_-_77279395 | 0.09 |
ENSMUST00000159300.8
|
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr2_+_150940165 | 0.09 |
ENSMUST00000109888.2
|
Gm14147
|
predicted gene 14147 |
| chrX_-_111315519 | 0.09 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr2_-_37007795 | 0.09 |
ENSMUST00000213817.2
ENSMUST00000215927.2 |
Olfr362
|
olfactory receptor 362 |
| chr2_-_26184450 | 0.09 |
ENSMUST00000217256.2
ENSMUST00000227200.2 |
Ccdc187
|
coiled-coil domain containing 187 |
| chr11_-_59462639 | 0.09 |
ENSMUST00000071943.6
|
Olfr222
|
olfactory receptor 222 |
| chr2_+_120459593 | 0.08 |
ENSMUST00000180041.9
|
Stard9
|
START domain containing 9 |
| chr13_-_67454476 | 0.08 |
ENSMUST00000049705.8
|
Zfp457
|
zinc finger protein 457 |
| chr10_+_129072073 | 0.08 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chr2_+_85805557 | 0.08 |
ENSMUST00000082191.5
|
Olfr1029
|
olfactory receptor 1029 |
| chr11_-_61233647 | 0.08 |
ENSMUST00000134423.3
ENSMUST00000093029.9 |
Slc47a2
|
solute carrier family 47, member 2 |
| chr16_+_57369595 | 0.08 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr1_-_173301492 | 0.08 |
ENSMUST00000169797.2
|
Ifi206
|
interferon activated gene 206 |
| chr3_+_92864693 | 0.08 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr8_+_34006758 | 0.08 |
ENSMUST00000149399.8
|
Tex15
|
testis expressed gene 15 |
| chr4_-_118667141 | 0.07 |
ENSMUST00000084313.5
ENSMUST00000219094.2 |
Olfr1335
|
olfactory receptor 1335 |
| chr2_+_65499097 | 0.07 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr2_+_61634797 | 0.07 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr11_-_115310743 | 0.07 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr6_+_41510925 | 0.07 |
ENSMUST00000103285.2
|
Trbj1-2
|
T cell receptor beta joining 1-2 |
| chr6_+_67993691 | 0.07 |
ENSMUST00000103314.3
|
Igkv1-122
|
immunoglobulin kappa chain variable 1-122 |
| chr14_+_25979825 | 0.07 |
ENSMUST00000173580.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr10_-_108535970 | 0.07 |
ENSMUST00000218023.2
|
Gm5136
|
predicted gene 5136 |
| chr16_-_58459102 | 0.07 |
ENSMUST00000053249.9
|
Ftdc2
|
ferritin domain containing 2 |
| chr18_+_37888770 | 0.06 |
ENSMUST00000061279.10
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr9_+_19247753 | 0.06 |
ENSMUST00000215572.3
ENSMUST00000213344.2 |
Olfr845
|
olfactory receptor 845 |
| chr1_-_194859015 | 0.06 |
ENSMUST00000082321.9
ENSMUST00000195120.6 |
Cr2
|
complement receptor 2 |
| chr3_+_79252668 | 0.06 |
ENSMUST00000164216.6
ENSMUST00000199658.2 |
Gm17359
|
predicted gene, 17359 |
| chr6_+_41512010 | 0.06 |
ENSMUST00000103288.2
|
Trbj1-5
|
T cell receptor beta joining 1-5 |
| chr3_+_32490300 | 0.06 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr1_+_118104272 | 0.06 |
ENSMUST00000186264.2
|
Gm29106
|
predicted gene 29106 |
| chr6_+_57086439 | 0.05 |
ENSMUST00000228270.2
|
Vmn1r10
|
vomeronasal 1 receptor 10 |
| chr7_+_26896401 | 0.05 |
ENSMUST00000155931.2
|
BC024978
|
cDNA sequence BC024978 |
| chr9_+_19533591 | 0.05 |
ENSMUST00000215372.2
|
Zfp317
|
zinc finger protein 317 |
| chr7_+_86895996 | 0.05 |
ENSMUST00000068829.13
|
Nox4
|
NADPH oxidase 4 |
| chr6_+_68196928 | 0.05 |
ENSMUST00000103318.6
ENSMUST00000103319.3 |
Igkv2-112
|
immunoglobulin kappa variable 2-112 |
| chr2_+_85876205 | 0.05 |
ENSMUST00000213496.2
|
Olfr1034
|
olfactory receptor 1034 |
| chr18_+_31742565 | 0.04 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr6_-_69477770 | 0.04 |
ENSMUST00000197448.2
|
Igkv4-58
|
immunoglobulin kappa variable 4-58 |
| chr14_-_63221950 | 0.04 |
ENSMUST00000100493.3
|
Defb48
|
defensin beta 48 |
| chr2_-_89671899 | 0.04 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr9_+_18767923 | 0.04 |
ENSMUST00000058411.4
|
Olfr829
|
olfactory receptor 829 |
| chr8_-_41494890 | 0.04 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr5_-_109340065 | 0.04 |
ENSMUST00000053253.10
|
Vmn2r13
|
vomeronasal 2, receptor 13 |
| chr7_-_103427550 | 0.04 |
ENSMUST00000072513.4
|
Olfr68
|
olfactory receptor 68 |
| chr14_+_26414422 | 0.04 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr13_-_66375387 | 0.03 |
ENSMUST00000167981.3
|
Gm10772
|
predicted gene 10772 |
| chr5_+_9316097 | 0.03 |
ENSMUST00000134991.8
ENSMUST00000069538.14 ENSMUST00000115348.9 |
Elapor2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr16_+_48877762 | 0.03 |
ENSMUST00000168680.2
|
Myh15
|
myosin, heavy chain 15 |
| chr2_-_150097511 | 0.03 |
ENSMUST00000063463.6
|
Gm21994
|
predicted gene 21994 |
| chr11_-_59484115 | 0.03 |
ENSMUST00000215626.2
|
Olfr223
|
olfactory receptor 223 |
| chr10_-_95509251 | 0.03 |
ENSMUST00000218771.2
ENSMUST00000099328.2 |
Anapc15-ps
|
anaphase promoting complex C subunit 15, pseudogene |
| chr1_+_164103127 | 0.03 |
ENSMUST00000027867.7
|
Ccdc181
|
coiled-coil domain containing 181 |
| chr12_+_76464372 | 0.03 |
ENSMUST00000220187.2
|
Ppp1r36
|
protein phosphatase 1, regulatory subunit 36 |
| chr2_+_103254401 | 0.03 |
ENSMUST00000028609.14
|
Elf5
|
E74-like factor 5 |
| chr1_+_24216691 | 0.03 |
ENSMUST00000054588.15
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr4_+_99184137 | 0.03 |
ENSMUST00000094955.3
|
Gm12689
|
predicted gene 12689 |
| chr6_+_68098030 | 0.03 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr7_+_139827152 | 0.03 |
ENSMUST00000164583.8
ENSMUST00000093984.3 |
Scart2
|
scavenger receptor family member expressed on T cells 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.3 | 0.8 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.2 | 2.0 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 0.6 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.2 | 0.6 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.7 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 1.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.9 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 1.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 | 0.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.2 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.9 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 1.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.2 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 3.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.6 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 1.1 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 5.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.3 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 2.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.2 | 4.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.5 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 2.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.8 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.2 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.1 | 1.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.0 | 7.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 7.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 1.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 2.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |