Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
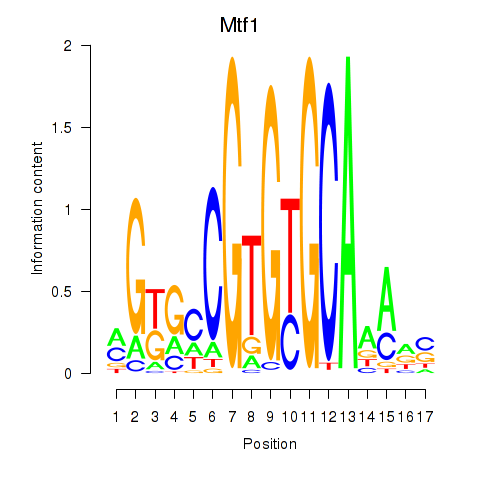
Results for Mtf1
Z-value: 2.63
Transcription factors associated with Mtf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mtf1
|
ENSMUSG00000028890.14 | Mtf1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mtf1 | mm39_v1_chr4_+_124696336_124696381 | 0.61 | 7.8e-05 | Click! |
Activity profile of Mtf1 motif
Sorted Z-values of Mtf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mtf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94899292 | 82.90 |
ENSMUST00000034214.8
ENSMUST00000212806.2 |
Mt2
|
metallothionein 2 |
| chr8_+_94905710 | 23.87 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr9_+_98372575 | 22.72 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr6_+_90596123 | 9.63 |
ENSMUST00000032177.10
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr3_+_107803225 | 9.28 |
ENSMUST00000172247.8
ENSMUST00000167387.8 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr3_+_107803137 | 8.95 |
ENSMUST00000004134.11
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr4_-_43040278 | 8.70 |
ENSMUST00000107958.8
ENSMUST00000107959.8 ENSMUST00000152846.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr3_+_107803563 | 8.68 |
ENSMUST00000169365.2
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr1_+_135656885 | 7.99 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr5_-_34794451 | 7.97 |
ENSMUST00000124668.2
ENSMUST00000001109.11 ENSMUST00000155577.8 ENSMUST00000114329.8 |
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr1_-_169358912 | 7.40 |
ENSMUST00000192248.2
ENSMUST00000028000.13 |
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr2_+_118943274 | 6.42 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr5_-_34794546 | 6.13 |
ENSMUST00000114331.10
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr17_-_71833752 | 5.65 |
ENSMUST00000232863.2
ENSMUST00000024851.10 |
Ndc80
|
NDC80 kinetochore complex component |
| chr1_-_169359015 | 5.52 |
ENSMUST00000111368.8
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr11_+_63023893 | 5.07 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chr12_-_36206780 | 4.64 |
ENSMUST00000223382.2
ENSMUST00000020856.6 |
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr9_-_66032134 | 4.59 |
ENSMUST00000034946.15
|
Snx1
|
sorting nexin 1 |
| chr3_-_121325887 | 4.40 |
ENSMUST00000039197.9
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr17_-_46513499 | 4.03 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr4_-_126057263 | 3.93 |
ENSMUST00000097891.4
|
Sh3d21
|
SH3 domain containing 21 |
| chr6_-_56900917 | 3.91 |
ENSMUST00000031793.8
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr11_+_63023395 | 3.82 |
ENSMUST00000108701.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr10_+_20828446 | 3.72 |
ENSMUST00000105525.12
|
Ahi1
|
Abelson helper integration site 1 |
| chr5_-_34794185 | 3.69 |
ENSMUST00000149657.5
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr8_+_71358576 | 3.60 |
ENSMUST00000019405.4
ENSMUST00000212511.2 |
Map1s
|
microtubule-associated protein 1S |
| chr1_+_75376714 | 3.45 |
ENSMUST00000113589.8
|
Speg
|
SPEG complex locus |
| chr12_-_36206750 | 3.41 |
ENSMUST00000221388.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_36206626 | 3.38 |
ENSMUST00000220828.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr15_-_83033508 | 3.12 |
ENSMUST00000100375.11
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr6_+_18848570 | 2.74 |
ENSMUST00000056398.11
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr12_+_111725282 | 2.74 |
ENSMUST00000239017.2
ENSMUST00000084941.12 |
Klc1
|
kinesin light chain 1 |
| chr11_-_32172233 | 2.70 |
ENSMUST00000150381.2
ENSMUST00000144902.2 ENSMUST00000020524.15 |
Rhbdf1
|
rhomboid 5 homolog 1 |
| chr15_-_83033559 | 2.67 |
ENSMUST00000058793.14
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr12_+_111725357 | 2.59 |
ENSMUST00000118471.8
ENSMUST00000122300.8 |
Klc1
|
kinesin light chain 1 |
| chr14_-_8123295 | 2.41 |
ENSMUST00000225775.2
ENSMUST00000224877.2 ENSMUST00000090591.4 |
Il3ra
|
interleukin 3 receptor, alpha chain |
| chr19_-_4665668 | 2.38 |
ENSMUST00000113822.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_+_58845502 | 2.32 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr19_-_4665509 | 2.31 |
ENSMUST00000053597.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr9_-_14292269 | 2.29 |
ENSMUST00000214236.2
|
Endod1
|
endonuclease domain containing 1 |
| chr5_-_100126707 | 2.21 |
ENSMUST00000170912.2
|
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr15_-_83033471 | 1.99 |
ENSMUST00000129372.2
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr4_-_126056412 | 1.89 |
ENSMUST00000094760.9
|
Sh3d21
|
SH3 domain containing 21 |
| chr8_+_93553901 | 1.67 |
ENSMUST00000034187.9
|
Mmp2
|
matrix metallopeptidase 2 |
| chr2_-_119617985 | 1.62 |
ENSMUST00000110793.8
ENSMUST00000099529.9 ENSMUST00000048493.12 |
Rpap1
|
RNA polymerase II associated protein 1 |
| chr10_+_17931459 | 1.61 |
ENSMUST00000154718.8
ENSMUST00000126390.8 ENSMUST00000164556.8 ENSMUST00000150029.8 |
Reps1
|
RalBP1 associated Eps domain containing protein |
| chr6_+_18848600 | 1.59 |
ENSMUST00000201141.3
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr1_+_128031055 | 1.55 |
ENSMUST00000188381.7
ENSMUST00000187900.7 ENSMUST00000036288.11 |
R3hdm1
|
R3H domain containing 1 |
| chr8_+_106937625 | 1.52 |
ENSMUST00000109297.8
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr7_-_119393182 | 1.50 |
ENSMUST00000106523.8
ENSMUST00000063902.14 ENSMUST00000150844.3 |
Eri2
|
exoribonuclease 2 |
| chr15_-_93417380 | 1.47 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr16_+_38405718 | 1.36 |
ENSMUST00000165631.2
|
Tmem39a
|
transmembrane protein 39a |
| chr4_+_135933676 | 1.33 |
ENSMUST00000047526.8
|
Asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr3_-_27764571 | 1.23 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr16_-_91394522 | 1.16 |
ENSMUST00000023686.15
|
Tmem50b
|
transmembrane protein 50B |
| chr8_+_106937568 | 1.15 |
ENSMUST00000071592.12
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr5_+_115644727 | 1.07 |
ENSMUST00000067268.15
ENSMUST00000086523.7 ENSMUST00000212819.3 |
Pxn
|
paxillin |
| chr9_+_19716202 | 1.05 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr7_-_42962640 | 0.94 |
ENSMUST00000012796.14
ENSMUST00000107986.9 |
Zfp715
|
zinc finger protein 715 |
| chr2_+_144210881 | 0.88 |
ENSMUST00000028911.15
ENSMUST00000147747.8 ENSMUST00000183618.3 |
Kat14
Pet117
|
lysine acetyltransferase 14 PET117 homolog |
| chr11_-_5848771 | 0.85 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr10_-_7162196 | 0.80 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3 |
| chr11_+_4845328 | 0.80 |
ENSMUST00000038237.8
|
Thoc5
|
THO complex 5 |
| chr7_+_119393210 | 0.80 |
ENSMUST00000033218.15
ENSMUST00000106520.9 |
Rexo5
|
RNA exonuclease 5 |
| chr16_-_18165876 | 0.79 |
ENSMUST00000125287.9
|
Tango2
|
transport and golgi organization 2 |
| chr11_+_4845314 | 0.78 |
ENSMUST00000101615.9
|
Thoc5
|
THO complex 5 |
| chr1_+_66426127 | 0.76 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr9_+_108368032 | 0.69 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr5_-_125418107 | 0.67 |
ENSMUST00000111390.8
ENSMUST00000086075.13 |
Scarb1
|
scavenger receptor class B, member 1 |
| chr2_+_28082943 | 0.67 |
ENSMUST00000113920.8
|
Olfm1
|
olfactomedin 1 |
| chr4_+_133302039 | 0.66 |
ENSMUST00000030662.3
|
Gpatch3
|
G patch domain containing 3 |
| chr4_+_152171286 | 0.57 |
ENSMUST00000118648.8
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr15_+_81686622 | 0.51 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr10_-_80837173 | 0.47 |
ENSMUST00000099462.8
ENSMUST00000118233.8 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr7_+_119393312 | 0.43 |
ENSMUST00000084644.3
|
Rexo5
|
RNA exonuclease 5 |
| chr15_+_27466732 | 0.36 |
ENSMUST00000022875.7
|
Ank
|
progressive ankylosis |
| chr14_+_59438658 | 0.34 |
ENSMUST00000173547.8
ENSMUST00000043227.13 ENSMUST00000022551.14 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr9_+_108367801 | 0.23 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr10_+_80972089 | 0.22 |
ENSMUST00000048128.15
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr5_+_31026967 | 0.20 |
ENSMUST00000114716.4
|
Tmem214
|
transmembrane protein 214 |
| chr5_-_140986312 | 0.18 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
| chr8_-_71229293 | 0.15 |
ENSMUST00000034296.15
|
Pik3r2
|
phosphoinositide-3-kinase regulatory subunit 2 |
| chr16_+_18166045 | 0.15 |
ENSMUST00000239533.1
ENSMUST00000239534.1 |
ARVCF
|
armadillo repeat deleted in velocardiofacial syndrome |
| chr4_+_138700195 | 0.10 |
ENSMUST00000123636.8
ENSMUST00000043042.10 ENSMUST00000050949.9 |
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr10_-_117074501 | 0.03 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 106.8 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 4.5 | 26.9 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 4.4 | 17.8 | GO:0015904 | tetracycline transport(GO:0015904) |
| 2.5 | 22.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 2.1 | 6.4 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 1.2 | 3.7 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 1.1 | 5.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.8 | 2.4 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.6 | 5.7 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.5 | 1.5 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.4 | 2.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.4 | 4.0 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.4 | 2.7 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.3 | 7.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 1.7 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.3 | 3.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 12.9 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.3 | 4.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 8.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.6 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.2 | 0.7 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.2 | 3.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 8.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.2 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.9 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 1.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 2.7 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 3.4 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.6 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 1.5 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 4.3 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 18.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.5 | 4.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.1 | 26.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 9.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.3 | 4.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.3 | 8.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 3.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 17.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 6.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 5.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 13.5 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 10.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 6.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.9 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.6 | GO:0030880 | RNA polymerase complex(GO:0030880) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.8 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 1.3 | 22.7 | GO:0019841 | retinol binding(GO:0019841) |
| 1.1 | 4.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.9 | 26.9 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.8 | 2.4 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-3 binding(GO:0019978) |
| 0.7 | 6.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 2.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 1.1 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.2 | 17.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 0.7 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 3.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 4.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 76.5 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 3.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 5.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 5.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 10.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 2.3 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 20.9 | GO:0003723 | RNA binding(GO:0003723) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 22.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 6.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 5.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 8.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 5.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 26.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 22.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 6.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 3.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 5.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 18.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 4.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |