Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
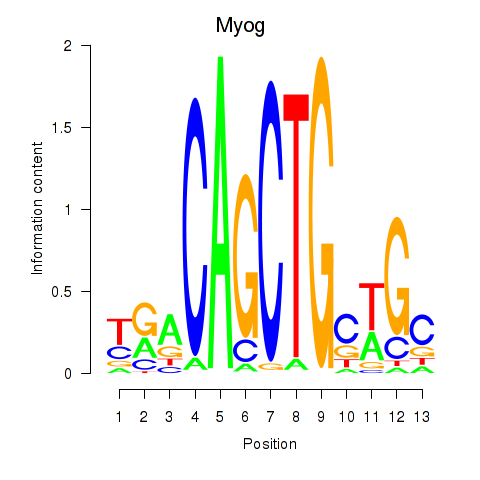
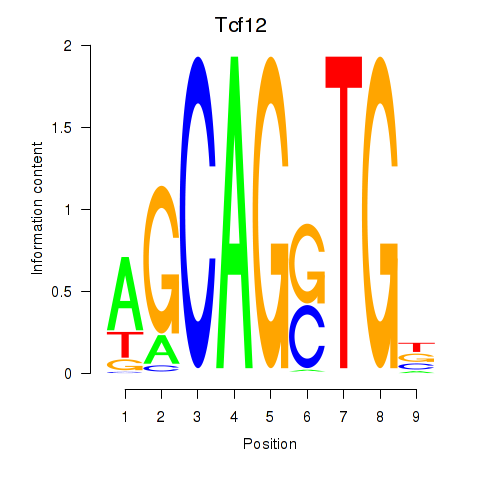
Results for Myog_Tcf12
Z-value: 2.23
Transcription factors associated with Myog_Tcf12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myog
|
ENSMUSG00000026459.6 | Myog |
|
Tcf12
|
ENSMUSG00000032228.17 | Tcf12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf12 | mm39_v1_chr9_-_72018933_72018993 | 0.46 | 4.8e-03 | Click! |
| Myog | mm39_v1_chr1_+_134217727_134217753 | 0.30 | 7.4e-02 | Click! |
Activity profile of Myog_Tcf12 motif
Sorted Z-values of Myog_Tcf12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Myog_Tcf12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142223662 | 15.25 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr11_+_99748741 | 14.69 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr6_+_29694181 | 13.35 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr7_+_44866635 | 12.44 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr1_-_75482975 | 11.04 |
ENSMUST00000113567.10
ENSMUST00000113565.3 |
Obsl1
|
obscurin-like 1 |
| chr11_-_102255999 | 10.73 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr12_+_109425769 | 9.82 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr18_+_34973605 | 7.95 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr1_-_132294807 | 7.79 |
ENSMUST00000136828.3
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr1_-_75110511 | 7.69 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr11_+_104468107 | 7.29 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr4_-_133694607 | 7.14 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_104467791 | 6.95 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr5_+_123214332 | 6.87 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr14_-_20319242 | 6.76 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr2_+_84564394 | 6.67 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr2_+_131028861 | 6.47 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr15_+_78128990 | 6.46 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr10_-_128236317 | 6.38 |
ENSMUST00000167859.2
ENSMUST00000218858.2 |
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_+_141995545 | 6.30 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr11_-_46203047 | 6.30 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr8_+_23629080 | 6.15 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr19_-_6996791 | 6.11 |
ENSMUST00000040772.9
|
Fermt3
|
fermitin family member 3 |
| chr4_-_133694543 | 6.03 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr7_+_142025817 | 5.81 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr7_+_142025575 | 5.73 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr12_-_76756772 | 5.72 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr13_-_55676334 | 5.68 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr10_-_128236366 | 5.62 |
ENSMUST00000219131.2
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr4_-_118314647 | 5.53 |
ENSMUST00000106375.2
ENSMUST00000168404.9 ENSMUST00000006556.11 |
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr7_+_126810780 | 5.52 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr15_-_78657640 | 5.49 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr15_+_78810919 | 5.42 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr11_-_99328969 | 5.34 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr4_+_137004793 | 5.34 |
ENSMUST00000045747.5
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr4_-_63965161 | 5.29 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr10_-_128237087 | 5.26 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_-_126303351 | 5.20 |
ENSMUST00000106364.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr3_-_98247237 | 5.19 |
ENSMUST00000065793.12
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr7_-_80037153 | 5.04 |
ENSMUST00000206728.2
|
Fes
|
feline sarcoma oncogene |
| chr4_-_133600308 | 4.98 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr4_-_118314707 | 4.94 |
ENSMUST00000102671.10
|
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr2_-_122441719 | 4.92 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chrX_-_149595711 | 4.91 |
ENSMUST00000112697.10
|
Maged2
|
MAGE family member D2 |
| chr4_-_43523388 | 4.89 |
ENSMUST00000107913.10
ENSMUST00000030184.12 |
Tpm2
|
tropomyosin 2, beta |
| chr19_-_6996696 | 4.87 |
ENSMUST00000236188.2
|
Fermt3
|
fermitin family member 3 |
| chr4_+_114916703 | 4.76 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr7_+_3339077 | 4.76 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_+_3339059 | 4.74 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr15_+_78129040 | 4.71 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr4_-_43523595 | 4.46 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr17_+_28988354 | 4.44 |
ENSMUST00000233109.2
ENSMUST00000004986.14 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr3_-_100396635 | 4.44 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr8_+_23629046 | 4.43 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr7_-_143013899 | 4.36 |
ENSMUST00000208137.2
ENSMUST00000207910.2 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chrX_-_149595873 | 4.24 |
ENSMUST00000131241.2
ENSMUST00000147152.3 ENSMUST00000143843.8 |
Maged2
|
MAGE family member D2 |
| chr7_+_44866095 | 4.15 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr10_+_60182630 | 4.08 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr15_+_73620213 | 4.06 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr11_+_115790768 | 4.03 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr4_-_131695135 | 4.01 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr8_-_25592001 | 3.97 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr19_-_24533183 | 3.84 |
ENSMUST00000112673.9
ENSMUST00000025800.15 |
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta |
| chr13_-_56326695 | 3.75 |
ENSMUST00000225063.2
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr17_+_28988271 | 3.75 |
ENSMUST00000233984.2
ENSMUST00000233460.2 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr11_+_69806866 | 3.73 |
ENSMUST00000134581.2
|
Gps2
|
G protein pathway suppressor 2 |
| chr16_-_16687119 | 3.69 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr10_+_128745214 | 3.62 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chrX_-_36253309 | 3.62 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chr5_+_97145533 | 3.60 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr3_-_95049655 | 3.59 |
ENSMUST00000013851.4
|
Tnfaip8l2
|
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr4_-_43523745 | 3.59 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chr13_-_56326511 | 3.57 |
ENSMUST00000169652.3
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr7_-_24705320 | 3.57 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr1_+_75336965 | 3.54 |
ENSMUST00000027409.10
|
Des
|
desmin |
| chr9_+_107852733 | 3.53 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr8_+_95703728 | 3.50 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr8_+_95721019 | 3.49 |
ENSMUST00000212976.2
ENSMUST00000212995.2 |
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_+_103061905 | 3.45 |
ENSMUST00000042286.12
ENSMUST00000218163.2 |
Fmnl1
|
formin-like 1 |
| chr7_-_44180700 | 3.43 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr4_-_133856025 | 3.38 |
ENSMUST00000105879.2
ENSMUST00000030651.9 |
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3 |
| chr11_-_69496655 | 3.38 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr5_-_114911548 | 3.38 |
ENSMUST00000178440.8
ENSMUST00000043283.14 ENSMUST00000112185.9 ENSMUST00000155908.8 |
Git2
|
GIT ArfGAP 2 |
| chr8_-_25592385 | 3.34 |
ENSMUST00000064883.14
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr7_-_126302315 | 3.34 |
ENSMUST00000173108.8
ENSMUST00000205515.2 |
Coro1a
|
coronin, actin binding protein 1A |
| chr17_-_43813664 | 3.32 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr4_-_116228921 | 3.31 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr11_-_107607343 | 3.29 |
ENSMUST00000021065.6
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr11_+_96820091 | 3.25 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr4_+_132968082 | 3.25 |
ENSMUST00000030677.7
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr5_-_138169509 | 3.24 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr15_-_36598263 | 3.22 |
ENSMUST00000155116.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_+_4204605 | 3.21 |
ENSMUST00000061086.9
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr12_-_113542610 | 3.21 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr19_+_5118103 | 3.18 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr2_-_93164812 | 3.17 |
ENSMUST00000111265.9
|
Tspan18
|
tetraspanin 18 |
| chr15_-_66703471 | 3.13 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr11_+_96820220 | 3.12 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr11_+_72851989 | 3.12 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr15_-_54953819 | 3.12 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr5_-_148989821 | 3.11 |
ENSMUST00000139443.8
ENSMUST00000085546.13 |
Hmgb1
|
high mobility group box 1 |
| chr17_-_26420300 | 3.06 |
ENSMUST00000025019.9
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr7_+_126811831 | 3.03 |
ENSMUST00000127710.3
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr5_-_148336711 | 2.91 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr16_+_87350202 | 2.91 |
ENSMUST00000026700.8
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr14_+_62529924 | 2.89 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chrX_-_73293425 | 2.89 |
ENSMUST00000114299.8
|
Flna
|
filamin, alpha |
| chr7_+_121888520 | 2.88 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr13_-_37233179 | 2.88 |
ENSMUST00000037491.11
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr9_-_21202545 | 2.85 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr6_-_91093766 | 2.83 |
ENSMUST00000113509.2
ENSMUST00000032179.14 |
Nup210
|
nucleoporin 210 |
| chr6_-_86646118 | 2.83 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr11_+_77384234 | 2.82 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chr11_-_94133527 | 2.82 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr17_-_26420332 | 2.82 |
ENSMUST00000121959.3
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr11_+_115790951 | 2.82 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr4_+_134195631 | 2.82 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr7_+_141996067 | 2.75 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr12_+_24758240 | 2.74 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr2_+_130119077 | 2.74 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr16_-_4831349 | 2.73 |
ENSMUST00000201077.2
ENSMUST00000202281.4 ENSMUST00000090453.9 ENSMUST00000023191.17 |
Rogdi
|
rogdi homolog |
| chr3_-_127689890 | 2.72 |
ENSMUST00000057198.9
ENSMUST00000199273.2 |
Fam241a
|
family with sequence similarity 241, member A |
| chr18_-_42084249 | 2.71 |
ENSMUST00000070949.6
ENSMUST00000235606.2 |
Prelid2
|
PRELI domain containing 2 |
| chr9_+_44245981 | 2.69 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr5_+_76331727 | 2.62 |
ENSMUST00000031144.14
|
Tmem165
|
transmembrane protein 165 |
| chr12_-_40087393 | 2.58 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr3_+_95496270 | 2.57 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr3_+_95496239 | 2.57 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr1_-_182929025 | 2.56 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr4_-_133599616 | 2.56 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_+_115705550 | 2.53 |
ENSMUST00000021134.10
ENSMUST00000106481.9 |
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chrX_-_48886577 | 2.53 |
ENSMUST00000033442.14
ENSMUST00000114891.2 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr5_+_21748523 | 2.52 |
ENSMUST00000035651.6
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr17_+_56610321 | 2.52 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr2_+_32253692 | 2.49 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr5_+_137744228 | 2.47 |
ENSMUST00000100539.10
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr17_+_47816042 | 2.47 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr5_+_137743992 | 2.46 |
ENSMUST00000100540.10
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr5_-_140634773 | 2.45 |
ENSMUST00000197452.5
ENSMUST00000042661.8 |
Ttyh3
|
tweety family member 3 |
| chr5_+_137756407 | 2.44 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr11_+_121150798 | 2.43 |
ENSMUST00000106113.2
|
Foxk2
|
forkhead box K2 |
| chr7_-_80037622 | 2.43 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr2_+_103800553 | 2.41 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr4_-_156340276 | 2.37 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr17_+_47906985 | 2.37 |
ENSMUST00000182539.8
|
Ccnd3
|
cyclin D3 |
| chr11_+_95227836 | 2.35 |
ENSMUST00000037502.7
|
Fam117a
|
family with sequence similarity 117, member A |
| chr17_+_28426752 | 2.35 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr17_+_47815968 | 2.35 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr10_-_80413119 | 2.35 |
ENSMUST00000038558.9
|
Klf16
|
Kruppel-like factor 16 |
| chr17_+_47816074 | 2.35 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr8_+_106245368 | 2.32 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr4_-_93223746 | 2.31 |
ENSMUST00000066774.6
|
Tusc1
|
tumor suppressor candidate 1 |
| chr12_+_108572015 | 2.30 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr5_-_114911509 | 2.30 |
ENSMUST00000086564.11
|
Git2
|
GIT ArfGAP 2 |
| chr9_-_57743989 | 2.29 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr5_-_31102829 | 2.29 |
ENSMUST00000031051.8
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr11_+_69856222 | 2.27 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr5_-_113968483 | 2.27 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr2_+_103800459 | 2.27 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr12_+_26519203 | 2.26 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr5_-_138170077 | 2.25 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr1_+_87603952 | 2.25 |
ENSMUST00000170300.8
ENSMUST00000167032.2 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr11_+_53410697 | 2.23 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr7_+_18817767 | 2.23 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr10_-_128755127 | 2.22 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr12_-_113561594 | 2.22 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr8_-_123425805 | 2.21 |
ENSMUST00000127984.9
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr17_+_56610396 | 2.20 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr11_+_51858476 | 2.18 |
ENSMUST00000102763.5
|
Cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr17_-_48235325 | 2.17 |
ENSMUST00000113263.8
ENSMUST00000097311.9 |
Foxp4
|
forkhead box P4 |
| chr17_+_34808772 | 2.17 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr6_-_125471666 | 2.17 |
ENSMUST00000032492.9
|
Cd9
|
CD9 antigen |
| chr3_+_95136248 | 2.17 |
ENSMUST00000107201.9
|
Cdc42se1
|
CDC42 small effector 1 |
| chr17_+_27775637 | 2.16 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_47816137 | 2.16 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr1_-_192880260 | 2.16 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr8_+_121264161 | 2.15 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr10_+_82821304 | 2.15 |
ENSMUST00000040110.8
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr9_+_59563838 | 2.14 |
ENSMUST00000163694.4
|
Pkm
|
pyruvate kinase, muscle |
| chr2_+_120307390 | 2.14 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr2_-_25086810 | 2.14 |
ENSMUST00000081869.7
|
Tor4a
|
torsin family 4, member A |
| chr16_-_18445172 | 2.14 |
ENSMUST00000231335.2
ENSMUST00000232653.2 |
Gm49601
Septin5
|
predicted gene, 49601 septin 5 |
| chr17_+_27775471 | 2.13 |
ENSMUST00000118599.9
ENSMUST00000232265.2 ENSMUST00000232013.2 ENSMUST00000114888.11 ENSMUST00000231874.2 ENSMUST00000119486.9 ENSMUST00000231825.2 ENSMUST00000231866.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_27775613 | 2.12 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_48607405 | 2.10 |
ENSMUST00000170941.3
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr9_+_59563872 | 2.10 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr6_-_68713748 | 2.09 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr8_+_106412905 | 2.09 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr5_-_114911432 | 2.09 |
ENSMUST00000112183.8
|
Git2
|
GIT ArfGAP 2 |
| chr12_-_36092475 | 2.09 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr12_+_24758724 | 2.08 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr19_+_32463151 | 2.08 |
ENSMUST00000025827.10
|
Minpp1
|
multiple inositol polyphosphate histidine phosphatase 1 |
| chr4_+_130643260 | 2.07 |
ENSMUST00000030316.7
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr5_-_107873883 | 2.07 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr1_+_180762587 | 2.05 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr10_-_62067026 | 2.05 |
ENSMUST00000047883.11
|
Tspan15
|
tetraspanin 15 |
| chr1_+_75483721 | 2.03 |
ENSMUST00000037330.5
|
Inha
|
inhibin alpha |
| chr17_+_48606948 | 2.03 |
ENSMUST00000233092.2
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr7_-_141023199 | 2.02 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr11_+_70323452 | 2.02 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 3.5 | 17.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.8 | 5.4 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.8 | 5.3 | GO:0061184 | positive regulation of dermatome development(GO:0061184) negative regulation of male gonad development(GO:2000019) |
| 1.6 | 4.9 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.6 | 4.8 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.5 | 7.3 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 1.4 | 11.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 1.3 | 8.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.3 | 5.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.3 | 10.2 | GO:0032796 | uropod organization(GO:0032796) |
| 1.2 | 9.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.1 | 9.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.0 | 3.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 1.0 | 5.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.0 | 2.9 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.9 | 2.6 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.8 | 13.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.8 | 7.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.8 | 2.3 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.8 | 2.3 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.7 | 2.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.7 | 2.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.7 | 3.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.7 | 7.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.7 | 16.7 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.7 | 2.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.6 | 5.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.6 | 2.5 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.6 | 3.6 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.6 | 7.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 2.9 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.6 | 4.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.6 | 1.7 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 | 2.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.5 | 0.5 | GO:0061183 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.5 | 1.6 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.5 | 2.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.5 | 2.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.5 | 5.9 | GO:1901249 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.5 | 2.6 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.5 | 2.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.5 | 5.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 1.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.5 | 3.5 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.5 | 8.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.5 | 3.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.5 | 2.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.5 | 1.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.5 | 4.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.5 | 1.4 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 0.5 | 11.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.5 | 3.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 1.8 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.5 | 2.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.5 | 2.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.5 | 9.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 3.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 1.3 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.4 | 4.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 13.1 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.4 | 0.8 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.4 | 2.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.4 | 2.8 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 1.2 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.4 | 1.6 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 4.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.4 | 1.2 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.4 | 7.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 1.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.4 | 1.5 | GO:1904008 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.4 | 10.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.4 | 4.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 2.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.4 | 3.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.4 | 1.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 5.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.3 | 4.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 2.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 4.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 1.0 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.3 | 1.0 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.3 | 2.0 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.3 | 1.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 | 5.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.3 | 1.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.3 | 14.8 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.3 | 4.8 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.3 | 3.8 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.3 | 1.3 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.3 | 0.6 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.3 | 4.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 1.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.3 | 0.9 | GO:2000769 | positive regulation of NMDA glutamate receptor activity(GO:1904783) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.3 | 0.9 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.3 | 0.9 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.8 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 4.7 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.3 | 11.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.3 | 9.8 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.3 | 8.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 3.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 1.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.3 | 12.5 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.3 | 0.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 0.5 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.3 | 0.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 3.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 1.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.3 | 1.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 1.7 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 4.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 0.7 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 1.4 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.2 | 0.7 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 1.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.7 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 0.2 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) |
| 0.2 | 2.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 4.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.2 | 9.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.4 | GO:0048389 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) metanephric comma-shaped body morphogenesis(GO:0072278) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 2.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 3.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 0.6 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 2.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.0 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 0.6 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.2 | 3.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 2.8 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.2 | 0.8 | GO:0090345 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.2 | 1.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 1.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 0.8 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.2 | 1.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 0.9 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 1.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 0.7 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.2 | 0.7 | GO:0060754 | regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 1.8 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 | 0.4 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 0.7 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 | 0.7 | GO:0090094 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.2 | 0.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 6.9 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 3.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 2.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 0.5 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 1.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 0.7 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 0.5 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 0.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.8 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 0.6 | GO:0035936 | testosterone secretion(GO:0035936) negative regulation of steroid hormone secretion(GO:2000832) regulation of testosterone secretion(GO:2000843) |
| 0.2 | 1.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 0.6 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 3.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 0.5 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 1.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.8 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 7.5 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.2 | 0.8 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.8 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.2 | 1.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 1.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.4 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 1.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.5 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 | 0.6 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 | 0.6 | GO:0061296 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 2.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 2.4 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.1 | 1.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.8 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.8 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.4 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 1.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 2.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 1.9 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.1 | 0.4 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.1 | 1.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 1.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 1.4 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.2 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 16.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 1.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.4 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 5.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 2.7 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 | 1.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 1.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 2.3 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.5 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.5 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.6 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.6 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 4.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 5.2 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.4 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 1.0 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 1.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 1.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.9 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.8 | GO:0014841 | skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.5 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 2.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.5 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.1 | 0.5 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.7 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 1.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 1.6 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 5.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 2.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 2.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.8 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 3.4 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 0.7 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.2 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.5 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 2.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 0.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.6 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 1.0 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 2.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 6.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 1.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.9 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 0.3 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.1 | 1.9 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 2.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 2.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.5 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 0.4 | GO:0034088 | maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.6 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 0.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.5 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 3.9 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.1 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 4.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.5 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 5.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 2.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 1.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 1.9 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.5 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 1.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 1.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.4 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.4 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 1.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 1.5 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.2 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.3 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 1.0 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.3 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.5 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.9 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.8 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.5 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0042533 | tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 2.1 | GO:0022408 | negative regulation of cell-cell adhesion(GO:0022408) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 1.1 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 3.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.7 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.7 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.4 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.1 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.6 | 4.8 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.4 | 4.2 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 1.3 | 15.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.2 | 3.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 1.1 | 8.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.9 | 10.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.9 | 12.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.8 | 2.5 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.8 | 4.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 6.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.7 | 11.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.7 | 5.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.7 | 3.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.7 | 6.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 11.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 3.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.6 | 28.2 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 1.7 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.5 | 1.5 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.5 | 7.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.5 | 2.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 2.9 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 2.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.4 | 14.4 | GO:0031672 | A band(GO:0031672) |
| 0.4 | 4.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.4 | 2.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 6.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 1.5 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.4 | 1.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 2.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 1.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.3 | 2.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 0.9 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.3 | 1.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 0.9 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 2.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 1.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 6.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 12.4 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 3.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 1.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 0.8 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 5.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 5.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 11.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 1.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 28.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.2 | 1.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 1.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 2.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 16.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.6 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 1.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 4.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 6.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.7 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 2.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 8.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 5.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 2.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.3 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.1 | 4.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.6 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 6.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.0 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 2.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 2.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 7.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 7.8 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 6.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 9.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0030666 | endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.9 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 7.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 4.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 2.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 13.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 10.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.4 | 5.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.0 | 3.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.9 | 9.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.9 | 3.7 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.9 | 9.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.9 | 11.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.8 | 4.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 4.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.8 | 2.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.8 | 2.3 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.7 | 17.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.7 | 2.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.7 | 2.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.7 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.7 | 3.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.7 | 4.7 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.7 | 13.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.7 | 5.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 1.8 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.6 | 5.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.6 | 6.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 3.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.5 | 10.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 5.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 4.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.5 | 2.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.5 | 2.0 | GO:0038100 | nodal binding(GO:0038100) |
| 0.5 | 1.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.5 | 4.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.5 | 4.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.5 | 1.4 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.5 | 2.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.4 | 1.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 2.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.4 | 9.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 4.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 3.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 1.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 1.7 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.4 | 2.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 4.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 15.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 7.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 3.4 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 7.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.4 | 1.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.4 | 11.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 19.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 1.4 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.3 | 5.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.3 | 2.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 14.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 0.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 1.8 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.3 | 21.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 5.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 7.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 2.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 1.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 2.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 1.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 13.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 3.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 1.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 1.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 0.6 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 1.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 5.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 0.8 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.2 | 3.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 23.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 1.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.7 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 5.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 3.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 4.3 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.2 | 0.7 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.5 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 5.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 6.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 1.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.6 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.2 | 0.6 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.6 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 0.5 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.1 | 1.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 5.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.8 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 1.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 3.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 6.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 2.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 1.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.1 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 1.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.8 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 2.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.6 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.6 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 4.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 1.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 4.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 10.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.3 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.1 | 1.0 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 3.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.5 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.1 | 30.0 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 1.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.2 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.1 | 5.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.2 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.1 | 1.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.9 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.5 | GO:0048018 | receptor agonist activity(GO:0048018) endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 3.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 4.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 8.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0034186 | lipopolysaccharide receptor activity(GO:0001875) apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 4.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 2.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.5 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 3.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 7.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 5.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.7 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 3.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 6.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 2.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 13.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 5.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 18.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 15.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 10.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 8.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 8.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 4.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 8.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 9.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 6.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 14.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 7.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 6.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 8.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 5.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 3.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 5.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.6 | 17.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.6 | 40.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 6.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 7.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 2.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 8.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 6.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 4.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 7.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 9.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 20.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 13.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 15.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 3.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 4.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 11.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 2.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 9.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 2.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 9.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 3.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 1.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 1.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 3.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 8.7 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 5.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 7.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 4.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 5.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 4.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 0.7 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 10.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 3.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 3.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 7.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 2.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |