Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
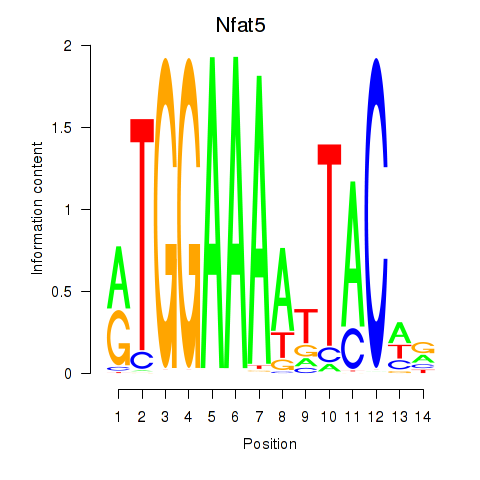
Results for Nfat5
Z-value: 0.54
Transcription factors associated with Nfat5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfat5
|
ENSMUSG00000003847.17 | Nfat5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfat5 | mm39_v1_chr8_+_108020092_108020118 | 0.37 | 2.6e-02 | Click! |
Activity profile of Nfat5 motif
Sorted Z-values of Nfat5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfat5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_99383938 | 1.83 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr1_+_87983099 | 1.75 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr3_-_98631847 | 1.70 |
ENSMUST00000107021.8
ENSMUST00000107022.8 |
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr10_-_95251327 | 1.58 |
ENSMUST00000172070.8
ENSMUST00000150432.8 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr1_+_87983189 | 1.48 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr10_-_95251145 | 1.41 |
ENSMUST00000119917.2
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr5_-_104169696 | 1.28 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr5_-_104169785 | 1.24 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr1_+_24717711 | 1.04 |
ENSMUST00000191471.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr4_-_49506538 | 1.04 |
ENSMUST00000043056.9
|
Baat
|
bile acid-Coenzyme A: amino acid N-acyltransferase |
| chr10_-_78187524 | 1.03 |
ENSMUST00000166360.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr10_-_78187887 | 1.03 |
ENSMUST00000105388.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr1_+_24717722 | 1.01 |
ENSMUST00000186096.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr6_+_91661074 | 1.00 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr10_-_78188046 | 0.98 |
ENSMUST00000146899.2
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr10_-_78187545 | 0.94 |
ENSMUST00000105390.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr1_-_93373145 | 0.92 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr1_+_24717793 | 0.92 |
ENSMUST00000186190.2
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr10_-_125164399 | 0.90 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr11_-_88754543 | 0.81 |
ENSMUST00000107904.3
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr6_+_91661034 | 0.75 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr10_+_21870565 | 0.73 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_+_120649233 | 0.71 |
ENSMUST00000068326.14
ENSMUST00000111890.9 ENSMUST00000076051.12 ENSMUST00000147496.2 |
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr2_+_67948057 | 0.71 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr15_+_25933632 | 0.68 |
ENSMUST00000228327.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr14_+_5894220 | 0.66 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr19_+_25214322 | 0.62 |
ENSMUST00000049400.15
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr11_+_120382666 | 0.58 |
ENSMUST00000026899.4
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
| chr7_-_80052491 | 0.56 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_-_177567397 | 0.53 |
ENSMUST00000108934.9
ENSMUST00000081529.11 |
Zfp972
|
zinc finger protein 972 |
| chrX_-_153999440 | 0.52 |
ENSMUST00000026318.15
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr2_-_30305313 | 0.50 |
ENSMUST00000132981.9
ENSMUST00000129494.2 |
Crat
|
carnitine acetyltransferase |
| chr18_+_36693646 | 0.47 |
ENSMUST00000155329.9
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr10_+_21869776 | 0.44 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_-_29411857 | 0.44 |
ENSMUST00000092623.5
|
Rspo3
|
R-spondin 3 |
| chrX_-_153999333 | 0.43 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr18_+_36693024 | 0.43 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr3_-_27764571 | 0.42 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr4_+_19575128 | 0.40 |
ENSMUST00000108253.8
ENSMUST00000029888.4 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr3_-_27764522 | 0.39 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr7_-_141472218 | 0.39 |
ENSMUST00000151890.3
|
Tollip
|
toll interacting protein |
| chr11_-_118292678 | 0.39 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr14_-_4808744 | 0.38 |
ENSMUST00000022303.17
ENSMUST00000091471.12 |
Thrb
|
thyroid hormone receptor beta |
| chr15_+_99476935 | 0.36 |
ENSMUST00000023752.6
|
Aqp2
|
aquaporin 2 |
| chr15_-_75766321 | 0.36 |
ENSMUST00000023237.8
|
Naprt
|
nicotinate phosphoribosyltransferase |
| chr12_-_83643964 | 0.35 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr9_+_92339422 | 0.34 |
ENSMUST00000034941.9
|
Plscr4
|
phospholipid scramblase 4 |
| chr16_+_84571011 | 0.34 |
ENSMUST00000114195.8
|
Jam2
|
junction adhesion molecule 2 |
| chrX_+_138585728 | 0.33 |
ENSMUST00000096313.4
|
Tbc1d8b
|
TBC1 domain family, member 8B |
| chrX_-_16683578 | 0.33 |
ENSMUST00000040820.13
|
Maob
|
monoamine oxidase B |
| chr6_+_73225616 | 0.32 |
ENSMUST00000203632.2
|
Suclg1
|
succinate-CoA ligase, GDP-forming, alpha subunit |
| chr12_-_83643883 | 0.31 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr10_+_4561974 | 0.30 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr7_-_30298287 | 0.30 |
ENSMUST00000108150.2
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr4_-_82423985 | 0.29 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr2_-_30305472 | 0.28 |
ENSMUST00000134120.2
ENSMUST00000102854.10 |
Crat
|
carnitine acetyltransferase |
| chr11_+_118913788 | 0.26 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr6_-_101354858 | 0.26 |
ENSMUST00000075994.11
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr11_-_118292758 | 0.25 |
ENSMUST00000043722.10
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr6_-_144155197 | 0.24 |
ENSMUST00000038815.14
ENSMUST00000111749.8 ENSMUST00000170367.9 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr12_+_98886826 | 0.24 |
ENSMUST00000085109.10
ENSMUST00000079146.13 |
Ttc8
|
tetratricopeptide repeat domain 8 |
| chr10_+_84458143 | 0.24 |
ENSMUST00000077175.7
|
Polr3b
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr4_-_82423944 | 0.23 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr15_-_38079089 | 0.23 |
ENSMUST00000110336.4
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr19_-_10502468 | 0.22 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr19_+_56414114 | 0.20 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr1_+_88334678 | 0.20 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr7_-_97066937 | 0.19 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr1_+_192984278 | 0.19 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chr11_+_83741657 | 0.19 |
ENSMUST00000021016.10
|
Hnf1b
|
HNF1 homeobox B |
| chr19_+_10502612 | 0.13 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr11_+_83741689 | 0.13 |
ENSMUST00000108114.9
|
Hnf1b
|
HNF1 homeobox B |
| chr5_-_66775979 | 0.13 |
ENSMUST00000162382.8
ENSMUST00000159786.8 ENSMUST00000162349.8 ENSMUST00000087256.12 ENSMUST00000160103.8 ENSMUST00000160870.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr7_-_126224848 | 0.11 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr19_-_10502546 | 0.11 |
ENSMUST00000237827.2
|
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr9_-_105398346 | 0.10 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr16_+_27208660 | 0.10 |
ENSMUST00000143823.2
|
Ccdc50
|
coiled-coil domain containing 50 |
| chr14_-_78866714 | 0.10 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr2_-_30305401 | 0.09 |
ENSMUST00000142096.2
|
Crat
|
carnitine acetyltransferase |
| chr9_+_7764042 | 0.09 |
ENSMUST00000052865.16
|
Tmem123
|
transmembrane protein 123 |
| chr5_-_69749617 | 0.09 |
ENSMUST00000173927.8
ENSMUST00000120789.8 ENSMUST00000031117.13 |
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr19_+_10502679 | 0.07 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr8_+_126456710 | 0.07 |
ENSMUST00000143504.8
|
Ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr8_-_70426910 | 0.07 |
ENSMUST00000116463.4
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr16_+_84571249 | 0.06 |
ENSMUST00000098407.3
|
Jam2
|
junction adhesion molecule 2 |
| chr6_-_144155167 | 0.06 |
ENSMUST00000077160.12
|
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr1_+_165312738 | 0.05 |
ENSMUST00000111440.8
ENSMUST00000027852.15 ENSMUST00000111439.8 |
Adcy10
|
adenylate cyclase 10 |
| chr6_+_67701864 | 0.03 |
ENSMUST00000103304.3
|
Igkv1-133
|
immunoglobulin kappa variable 1-133 |
| chr2_-_11506511 | 0.03 |
ENSMUST00000183869.8
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr18_-_78640066 | 0.03 |
ENSMUST00000235389.2
ENSMUST00000237674.2 |
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chrX_-_17437801 | 0.03 |
ENSMUST00000177213.8
|
Fundc1
|
FUN14 domain containing 1 |
| chr2_-_90054837 | 0.02 |
ENSMUST00000213994.3
|
Olfr1506
|
olfactory receptor 1506 |
| chr8_-_25581354 | 0.02 |
ENSMUST00000125466.2
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chrX_+_37861548 | 0.02 |
ENSMUST00000050744.6
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr14_+_59716265 | 0.02 |
ENSMUST00000224893.2
|
Cab39l
|
calcium binding protein 39-like |
| chr7_+_140658101 | 0.01 |
ENSMUST00000106039.9
|
Pkp3
|
plakophilin 3 |
| chr18_+_61408073 | 0.01 |
ENSMUST00000135688.2
|
Pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr7_+_108549545 | 0.01 |
ENSMUST00000207583.2
|
Tub
|
tubby bipartite transcription factor |
| chr11_+_97206542 | 0.01 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr13_+_23343482 | 0.01 |
ENSMUST00000226845.2
ENSMUST00000228666.2 ENSMUST00000227388.2 |
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chr9_-_29874401 | 0.00 |
ENSMUST00000075069.11
|
Ntm
|
neurotrimin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.6 | 2.5 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.6 | 1.8 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.3 | 3.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 1.0 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:0015744 | succinate transport(GO:0015744) |
| 0.1 | 0.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.0 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 0.9 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.4 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.8 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 3.0 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.3 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.3 | GO:0061228 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 1.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 0.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.2 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.1 | 0.8 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 3.6 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 3.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0005368 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.5 | 3.0 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 3.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.7 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 0.9 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.2 | 0.6 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 0.7 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.2 | 0.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 4.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.5 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 1.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.9 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.3 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 0.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 3.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 4.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |