Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
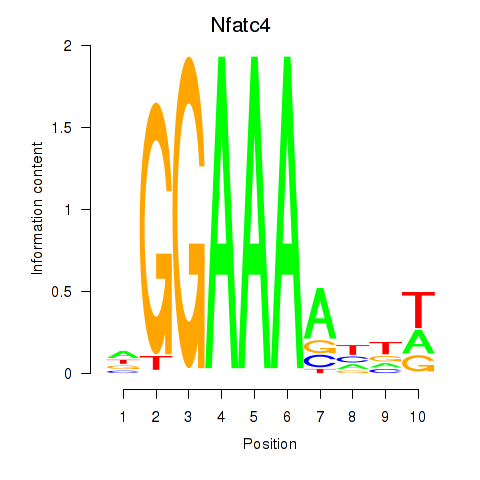
Results for Nfatc4
Z-value: 0.91
Transcription factors associated with Nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc4
|
ENSMUSG00000023411.13 | Nfatc4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc4 | mm39_v1_chr14_+_56062252_56062348 | -0.67 | 9.2e-06 | Click! |
Activity profile of Nfatc4 motif
Sorted Z-values of Nfatc4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_150341911 | 5.10 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr12_-_84497718 | 4.37 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr11_+_75358866 | 3.42 |
ENSMUST00000043598.14
ENSMUST00000108435.2 |
Tlcd2
|
TLC domain containing 2 |
| chr8_+_129085719 | 2.33 |
ENSMUST00000026917.10
|
Nrp1
|
neuropilin 1 |
| chr3_-_20329823 | 2.10 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr6_-_21851827 | 2.09 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr15_+_3300249 | 1.99 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr14_-_25927674 | 1.92 |
ENSMUST00000100811.6
|
Tmem254a
|
transmembrane protein 254a |
| chr14_-_25928096 | 1.79 |
ENSMUST00000185006.9
|
Tmem254a
|
transmembrane protein 254a |
| chr10_-_53952686 | 1.77 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr13_-_53135064 | 1.59 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr12_+_8027767 | 1.52 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr11_-_35871300 | 1.45 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr19_-_4993060 | 1.33 |
ENSMUST00000133504.2
ENSMUST00000133254.2 ENSMUST00000120475.8 ENSMUST00000025834.15 |
Peli3
|
pellino 3 |
| chr19_+_8816663 | 1.28 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr13_+_25127127 | 1.18 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr14_+_53980561 | 1.07 |
ENSMUST00000103667.6
|
Trav16
|
T cell receptor alpha variable 16 |
| chr10_+_4561974 | 0.90 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr15_-_79816785 | 0.84 |
ENSMUST00000089293.11
ENSMUST00000109616.9 |
Cbx7
|
chromobox 7 |
| chr14_-_65187287 | 0.81 |
ENSMUST00000067843.10
ENSMUST00000176489.8 ENSMUST00000175905.8 ENSMUST00000022544.14 ENSMUST00000175744.8 ENSMUST00000176128.8 |
Hmbox1
|
homeobox containing 1 |
| chr4_-_148244299 | 0.78 |
ENSMUST00000151127.8
ENSMUST00000105705.9 |
Fbxo44
|
F-box protein 44 |
| chr10_-_12689345 | 0.77 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr15_-_79816717 | 0.71 |
ENSMUST00000177044.2
ENSMUST00000109615.8 |
Cbx7
|
chromobox 7 |
| chr11_-_99412084 | 0.71 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chrX_+_7504913 | 0.70 |
ENSMUST00000128890.2
|
Syp
|
synaptophysin |
| chr12_+_84498196 | 0.70 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr11_-_99412162 | 0.59 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr8_-_39128662 | 0.55 |
ENSMUST00000118896.2
|
Sgcz
|
sarcoglycan zeta |
| chr12_+_76580386 | 0.54 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr1_+_174257996 | 0.53 |
ENSMUST00000053178.5
|
Olfr414
|
olfactory receptor 414 |
| chr2_-_73490719 | 0.50 |
ENSMUST00000154258.8
|
Chn1
|
chimerin 1 |
| chr2_+_124978518 | 0.50 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr4_+_130001349 | 0.48 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr14_+_70793340 | 0.46 |
ENSMUST00000163060.2
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr17_+_48037758 | 0.45 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr4_-_3938352 | 0.44 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr14_+_49409659 | 0.41 |
ENSMUST00000153488.9
|
Naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr9_+_51959415 | 0.39 |
ENSMUST00000000590.16
ENSMUST00000238858.2 |
Rdx
|
radixin |
| chr9_+_51959534 | 0.38 |
ENSMUST00000061352.11
|
Rdx
|
radixin |
| chr16_+_43184507 | 0.36 |
ENSMUST00000148775.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr12_+_52550775 | 0.35 |
ENSMUST00000219443.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr2_+_124978612 | 0.34 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr19_+_8828132 | 0.33 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr8_-_68270936 | 0.33 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_168259710 | 0.32 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr2_+_55325931 | 0.31 |
ENSMUST00000067101.10
|
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr9_-_64858878 | 0.30 |
ENSMUST00000037798.8
|
Slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr5_-_112542671 | 0.30 |
ENSMUST00000196256.2
|
Asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr8_-_41494890 | 0.30 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr1_-_169938298 | 0.30 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr10_-_44334711 | 0.29 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr11_-_89587671 | 0.28 |
ENSMUST00000128717.9
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr11_-_47270201 | 0.27 |
ENSMUST00000077221.6
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr12_+_103564479 | 0.27 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr10_+_33071964 | 0.27 |
ENSMUST00000219211.2
|
Trdn
|
triadin |
| chr19_+_26600820 | 0.25 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_68270870 | 0.23 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr3_-_37180093 | 0.23 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr6_-_128599779 | 0.23 |
ENSMUST00000203275.3
|
Klrb1a
|
killer cell lectin-like receptor subfamily B member 1A |
| chrX_+_56144946 | 0.22 |
ENSMUST00000141936.2
|
Vgll1
|
vestigial like family member 1 |
| chr10_+_23727325 | 0.20 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr3_-_105908789 | 0.20 |
ENSMUST00000066319.8
|
Pifo
|
primary cilia formation |
| chr14_+_120513106 | 0.19 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr10_-_44334683 | 0.19 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr3_+_94744844 | 0.19 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr14_+_120513076 | 0.17 |
ENSMUST00000088419.13
|
Mbnl2
|
muscleblind like splicing factor 2 |
| chr1_-_144052997 | 0.15 |
ENSMUST00000111941.2
ENSMUST00000052375.8 |
Rgs13
|
regulator of G-protein signaling 13 |
| chr1_+_34044940 | 0.13 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr1_+_82891043 | 0.13 |
ENSMUST00000220768.2
|
A030005L19Rik
|
RIKEN cDNA A030005L19 gene |
| chr10_-_89270527 | 0.13 |
ENSMUST00000218764.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr9_+_88838953 | 0.11 |
ENSMUST00000098485.4
|
Bcl2a1a
|
B cell leukemia/lymphoma 2 related protein A1a |
| chr9_-_13358049 | 0.11 |
ENSMUST00000167906.3
|
Gm17571
|
predicted gene, 17571 |
| chr5_+_53747796 | 0.11 |
ENSMUST00000113865.5
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr17_+_34129221 | 0.10 |
ENSMUST00000174541.9
|
Daxx
|
Fas death domain-associated protein |
| chr5_+_117501557 | 0.10 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr18_+_42186757 | 0.10 |
ENSMUST00000074679.4
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr11_-_53509485 | 0.10 |
ENSMUST00000000889.7
|
Il4
|
interleukin 4 |
| chr1_+_78794475 | 0.09 |
ENSMUST00000057262.8
ENSMUST00000187432.2 |
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr18_+_42186713 | 0.09 |
ENSMUST00000072008.11
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr6_+_21986445 | 0.08 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr3_-_86455575 | 0.08 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr10_+_21869776 | 0.07 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_+_48049080 | 0.06 |
ENSMUST00000153369.2
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr7_+_91321500 | 0.05 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr9_+_77543776 | 0.05 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr18_+_73996743 | 0.04 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr1_-_169938060 | 0.04 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr12_+_91367764 | 0.04 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr2_+_61876806 | 0.01 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.8 | 2.3 | GO:0097374 | cell migration involved in vasculogenesis(GO:0035441) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) sensory neuron axon guidance(GO:0097374) |
| 0.6 | 5.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.3 | 1.2 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 2.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 0.7 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 | 0.5 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 0.5 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.2 | 0.5 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.9 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 1.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.7 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 0.8 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 1.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.8 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.3 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.1 | 1.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.3 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 1.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.0 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.0 | 0.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.4 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:1900625 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.3 | 2.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.3 | 0.9 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 5.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 2.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.8 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 4.4 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 5.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 4.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |