Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
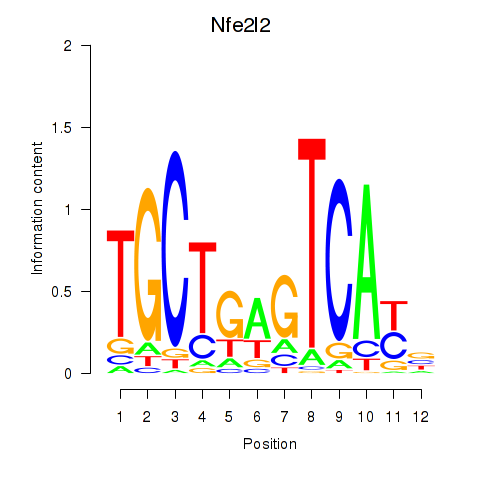
Results for Nfe2l2
Z-value: 1.06
Transcription factors associated with Nfe2l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l2
|
ENSMUSG00000015839.7 | Nfe2l2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2l2 | mm39_v1_chr2_-_75534985_75535023 | -0.24 | 1.6e-01 | Click! |
Activity profile of Nfe2l2 motif
Sorted Z-values of Nfe2l2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_86577940 | 5.50 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr19_-_4087907 | 4.98 |
ENSMUST00000237982.2
|
Gstp1
|
glutathione S-transferase, pi 1 |
| chr19_-_4087940 | 4.94 |
ENSMUST00000237893.2
ENSMUST00000169613.4 |
Gstp1
|
glutathione S-transferase, pi 1 |
| chr12_-_103925197 | 4.51 |
ENSMUST00000122229.8
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr19_-_4092218 | 4.43 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr16_+_22738987 | 3.81 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr1_+_58069090 | 3.64 |
ENSMUST00000001027.7
|
Aox1
|
aldehyde oxidase 1 |
| chr16_+_22739191 | 3.28 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr13_-_42001075 | 3.23 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr1_-_121260298 | 3.08 |
ENSMUST00000071064.13
|
Insig2
|
insulin induced gene 2 |
| chr5_-_24963006 | 3.03 |
ENSMUST00000047119.5
|
Crygn
|
crystallin, gamma N |
| chr3_+_146302832 | 2.94 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr9_-_121745354 | 2.94 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr13_-_42001102 | 2.93 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr15_-_82291372 | 2.89 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr13_-_42000958 | 2.79 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr6_+_129510331 | 2.30 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr16_+_90017634 | 2.18 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr9_-_106353792 | 2.16 |
ENSMUST00000214682.2
ENSMUST00000112479.9 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr11_+_120563844 | 2.16 |
ENSMUST00000106158.9
ENSMUST00000103016.8 ENSMUST00000168714.9 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr7_+_127399776 | 2.12 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_-_121260274 | 2.05 |
ENSMUST00000161068.2
|
Insig2
|
insulin induced gene 2 |
| chr11_-_50101592 | 2.00 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chr7_+_127400016 | 1.93 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr9_-_106353571 | 1.92 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr5_-_45607485 | 1.92 |
ENSMUST00000154962.8
ENSMUST00000118097.8 ENSMUST00000198258.5 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr13_-_12476313 | 1.90 |
ENSMUST00000143693.8
ENSMUST00000144283.2 ENSMUST00000099820.10 ENSMUST00000135166.8 |
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr11_+_120564185 | 1.89 |
ENSMUST00000135346.8
ENSMUST00000127269.8 ENSMUST00000131727.9 ENSMUST00000149389.8 ENSMUST00000153346.8 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr7_+_127399789 | 1.89 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr5_-_45607554 | 1.89 |
ENSMUST00000015950.12
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr18_-_3281089 | 1.88 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr5_-_45607463 | 1.87 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr11_-_69696428 | 1.78 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr19_-_4889284 | 1.59 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr4_+_116542741 | 1.54 |
ENSMUST00000135573.8
ENSMUST00000151129.8 |
Prdx1
|
peroxiredoxin 1 |
| chr19_-_4889314 | 1.53 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr11_+_80319424 | 1.46 |
ENSMUST00000173938.8
ENSMUST00000017572.14 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr5_-_108823435 | 1.43 |
ENSMUST00000051757.14
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr18_+_12637217 | 1.42 |
ENSMUST00000188815.2
|
Lama3
|
laminin, alpha 3 |
| chr7_+_127399848 | 1.42 |
ENSMUST00000139068.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr4_+_116543045 | 1.40 |
ENSMUST00000129315.8
ENSMUST00000106470.8 |
Prdx1
|
peroxiredoxin 1 |
| chr8_+_106052970 | 1.39 |
ENSMUST00000015000.12
ENSMUST00000098453.9 |
Tmem208
|
transmembrane protein 208 |
| chr1_+_165596961 | 1.36 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr4_+_117109204 | 1.29 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr4_+_117109148 | 1.28 |
ENSMUST00000062824.12
|
Tmem53
|
transmembrane protein 53 |
| chr8_+_120163857 | 1.27 |
ENSMUST00000152420.8
ENSMUST00000212112.2 ENSMUST00000098365.4 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr11_-_50216426 | 1.20 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr13_-_23894828 | 1.20 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr2_-_163261439 | 1.19 |
ENSMUST00000046908.10
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr3_-_116762617 | 1.18 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chrX_+_5959507 | 1.17 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr2_+_25285878 | 1.17 |
ENSMUST00000028328.3
|
Entpd2
|
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr15_-_76501525 | 1.16 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr7_-_30810422 | 1.14 |
ENSMUST00000039435.15
|
Hpn
|
hepsin |
| chr17_-_24863956 | 1.14 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr2_+_155617251 | 1.13 |
ENSMUST00000029141.6
|
Mmp24
|
matrix metallopeptidase 24 |
| chr5_+_31274046 | 1.12 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr14_+_4230569 | 1.12 |
ENSMUST00000090543.6
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr11_-_98329782 | 1.11 |
ENSMUST00000002655.8
|
Mien1
|
migration and invasion enhancer 1 |
| chr15_-_75886166 | 1.08 |
ENSMUST00000060807.12
|
Fam83h
|
family with sequence similarity 83, member H |
| chr5_-_147831610 | 1.06 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr2_+_25318642 | 1.05 |
ENSMUST00000102919.4
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr16_+_4825146 | 1.03 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr17_-_24863907 | 1.01 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr16_+_4825170 | 0.98 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr3_+_27237114 | 0.94 |
ENSMUST00000046515.15
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr14_+_28740162 | 0.92 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr2_+_151947444 | 0.91 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr15_+_54434576 | 0.90 |
ENSMUST00000025356.4
|
Mal2
|
mal, T cell differentiation protein 2 |
| chr13_-_64460491 | 0.89 |
ENSMUST00000222570.2
ENSMUST00000220895.2 |
Prxl2c
|
peroxiredoxin like 2C |
| chr4_+_122910382 | 0.87 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr10_+_127894816 | 0.85 |
ENSMUST00000052798.14
|
Ptges3
|
prostaglandin E synthase 3 |
| chr5_+_31274064 | 0.83 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr10_+_127894843 | 0.83 |
ENSMUST00000084771.3
|
Ptges3
|
prostaglandin E synthase 3 |
| chr16_+_20367327 | 0.82 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr14_-_30637344 | 0.80 |
ENSMUST00000226547.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr6_+_30509826 | 0.80 |
ENSMUST00000031797.11
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr1_+_107517726 | 0.79 |
ENSMUST00000000514.11
ENSMUST00000112706.4 |
Serpinb8
|
serine (or cysteine) peptidase inhibitor, clade B, member 8 |
| chr7_-_28879879 | 0.78 |
ENSMUST00000209034.2
ENSMUST00000182328.8 ENSMUST00000186182.2 ENSMUST00000059642.17 |
Psmd8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr12_-_83609217 | 0.73 |
ENSMUST00000222448.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr2_-_164621641 | 0.72 |
ENSMUST00000103095.5
|
Tnnc2
|
troponin C2, fast |
| chr2_+_11710101 | 0.71 |
ENSMUST00000138349.8
ENSMUST00000135341.8 ENSMUST00000128156.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr17_-_28569574 | 0.68 |
ENSMUST00000114799.8
ENSMUST00000219703.3 |
Tead3
|
TEA domain family member 3 |
| chr3_-_116762476 | 0.68 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr6_-_146403410 | 0.67 |
ENSMUST00000053273.15
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr1_-_172047282 | 0.66 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chr18_-_3280999 | 0.66 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr11_-_120328458 | 0.65 |
ENSMUST00000044271.15
ENSMUST00000103017.4 |
Nploc4
|
NPL4 homolog, ubiquitin recognition factor |
| chr18_-_35087355 | 0.64 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr16_-_17348882 | 0.64 |
ENSMUST00000231548.2
ENSMUST00000232041.2 ENSMUST00000231288.2 |
Thap7
|
THAP domain containing 7 |
| chr1_-_120001752 | 0.63 |
ENSMUST00000056089.8
|
Tmem37
|
transmembrane protein 37 |
| chr10_-_95158827 | 0.62 |
ENSMUST00000220279.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr4_-_57956411 | 0.62 |
ENSMUST00000030051.6
|
Txn1
|
thioredoxin 1 |
| chr7_-_103964662 | 0.59 |
ENSMUST00000106837.8
ENSMUST00000106839.9 ENSMUST00000070943.7 |
Trim12a
|
tripartite motif-containing 12A |
| chr19_+_8816663 | 0.56 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr10_-_34294461 | 0.53 |
ENSMUST00000213269.2
ENSMUST00000099973.4 ENSMUST00000105512.8 ENSMUST00000047885.14 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr3_-_141687987 | 0.53 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr10_+_60113449 | 0.52 |
ENSMUST00000105465.8
ENSMUST00000179238.8 ENSMUST00000177779.8 ENSMUST00000004316.15 |
Psap
|
prosaposin |
| chr12_+_55445560 | 0.52 |
ENSMUST00000021412.9
|
Psma6
|
proteasome subunit alpha 6 |
| chr7_-_18350423 | 0.51 |
ENSMUST00000057810.7
|
Psg23
|
pregnancy-specific glycoprotein 23 |
| chr6_-_146403638 | 0.50 |
ENSMUST00000079573.13
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr11_-_59700820 | 0.49 |
ENSMUST00000047706.3
ENSMUST00000102697.10 |
Flcn
|
folliculin |
| chr2_+_11710523 | 0.49 |
ENSMUST00000138856.2
ENSMUST00000078834.12 ENSMUST00000114834.10 ENSMUST00000114833.10 ENSMUST00000114831.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr2_+_11710246 | 0.49 |
ENSMUST00000148748.8
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr13_-_64460382 | 0.48 |
ENSMUST00000021938.11
ENSMUST00000221118.2 ENSMUST00000221350.2 |
Prxl2c
|
peroxiredoxin like 2C |
| chr2_+_112092271 | 0.48 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr12_+_71021395 | 0.47 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr15_-_79489286 | 0.44 |
ENSMUST00000229408.2
ENSMUST00000023065.8 |
Dmc1
|
DNA meiotic recombinase 1 |
| chr11_+_78068931 | 0.44 |
ENSMUST00000147819.8
|
Tlcd1
|
TLC domain containing 1 |
| chr7_+_44498415 | 0.43 |
ENSMUST00000107885.8
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr17_-_43003135 | 0.42 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr3_+_27237143 | 0.42 |
ENSMUST00000091284.5
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr11_+_121036969 | 0.42 |
ENSMUST00000039088.9
ENSMUST00000155694.2 |
Tex19.1
|
testis expressed gene 19.1 |
| chr14_-_77274056 | 0.41 |
ENSMUST00000062789.15
|
Lacc1
|
laccase domain containing 1 |
| chr1_-_155022501 | 0.41 |
ENSMUST00000027744.10
|
Mr1
|
major histocompatibility complex, class I-related |
| chr9_+_32135781 | 0.40 |
ENSMUST00000183121.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr6_-_40976413 | 0.40 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr1_+_33947250 | 0.39 |
ENSMUST00000183034.5
|
Dst
|
dystonin |
| chr14_+_4230658 | 0.38 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr13_-_58363431 | 0.38 |
ENSMUST00000076454.8
ENSMUST00000058735.12 |
Ubqln1
|
ubiquilin 1 |
| chr7_+_44984681 | 0.36 |
ENSMUST00000085351.7
|
Hrc
|
histidine rich calcium binding protein |
| chr7_+_139414057 | 0.35 |
ENSMUST00000026548.14
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr19_-_43928583 | 0.35 |
ENSMUST00000212396.2
|
Dnmbp
|
dynamin binding protein |
| chr15_+_80139371 | 0.34 |
ENSMUST00000109605.5
ENSMUST00000229828.2 |
Atf4
|
activating transcription factor 4 |
| chr13_-_65353757 | 0.34 |
ENSMUST00000223418.2
ENSMUST00000222559.2 ENSMUST00000221659.2 ENSMUST00000222273.2 |
Nlrp4f
|
NLR family, pyrin domain containing 4F |
| chr9_+_32305259 | 0.33 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr9_+_32135540 | 0.33 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_+_127511681 | 0.33 |
ENSMUST00000033070.9
|
Kat8
|
K(lysine) acetyltransferase 8 |
| chr11_-_121009503 | 0.32 |
ENSMUST00000039146.4
|
Tex19.2
|
testis expressed gene 19.2 |
| chr2_-_80411578 | 0.31 |
ENSMUST00000028386.12
|
Nckap1
|
NCK-associated protein 1 |
| chr12_-_76842263 | 0.31 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr6_+_124973752 | 0.30 |
ENSMUST00000162000.4
|
Pianp
|
PILR alpha associated neural protein |
| chr16_-_48592319 | 0.30 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr2_-_80411724 | 0.30 |
ENSMUST00000111760.3
|
Nckap1
|
NCK-associated protein 1 |
| chrX_+_72719098 | 0.29 |
ENSMUST00000171398.2
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr11_-_120358239 | 0.28 |
ENSMUST00000076921.7
|
Arl16
|
ADP-ribosylation factor-like 16 |
| chr6_+_124973644 | 0.28 |
ENSMUST00000032479.11
|
Pianp
|
PILR alpha associated neural protein |
| chr19_-_4927910 | 0.28 |
ENSMUST00000006626.5
|
Actn3
|
actinin alpha 3 |
| chr7_-_44646645 | 0.27 |
ENSMUST00000207342.2
|
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr3_+_137329433 | 0.26 |
ENSMUST00000053855.8
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chrX_-_110372800 | 0.25 |
ENSMUST00000137712.9
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr11_+_120358461 | 0.25 |
ENSMUST00000140862.7
ENSMUST00000106205.9 ENSMUST00000106203.9 |
Hgs
|
HGF-regulated tyrosine kinase substrate |
| chr11_+_4207557 | 0.25 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr16_-_48592372 | 0.25 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chrX_-_96420756 | 0.25 |
ENSMUST00000113832.2
ENSMUST00000037353.10 |
Eda2r
|
ectodysplasin A2 receptor |
| chr3_+_100732768 | 0.25 |
ENSMUST00000054791.9
|
Vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr16_+_17379749 | 0.24 |
ENSMUST00000171002.10
ENSMUST00000023441.11 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr14_-_21798678 | 0.24 |
ENSMUST00000075040.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chr5_+_143349645 | 0.24 |
ENSMUST00000010969.15
|
Grid2ip
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein 1 |
| chr2_+_11710633 | 0.24 |
ENSMUST00000114832.3
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr1_-_10790120 | 0.24 |
ENSMUST00000035577.7
|
Cpa6
|
carboxypeptidase A6 |
| chrX_+_52726793 | 0.23 |
ENSMUST00000069209.2
|
Ct55
|
cancer/testis antigen 55 |
| chr18_+_20380397 | 0.23 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr7_+_101875019 | 0.23 |
ENSMUST00000142873.8
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr14_-_20443676 | 0.23 |
ENSMUST00000061444.5
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chr15_-_101801351 | 0.23 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr19_+_4889394 | 0.23 |
ENSMUST00000088653.3
|
Ccdc87
|
coiled-coil domain containing 87 |
| chr16_-_16962256 | 0.22 |
ENSMUST00000115711.10
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr18_-_63825491 | 0.21 |
ENSMUST00000237004.2
|
Txnl1
|
thioredoxin-like 1 |
| chr12_-_85335193 | 0.21 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr18_+_20509771 | 0.21 |
ENSMUST00000076737.7
|
Dsg1b
|
desmoglein 1 beta |
| chr6_+_117988399 | 0.20 |
ENSMUST00000164960.4
|
Rasgef1a
|
RasGEF domain family, member 1A |
| chr9_-_65330231 | 0.19 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr7_+_26135039 | 0.19 |
ENSMUST00000119386.8
ENSMUST00000146907.3 |
Nlrp4a
|
NLR family, pyrin domain containing 4A |
| chr18_-_63825380 | 0.19 |
ENSMUST00000025476.4
|
Txnl1
|
thioredoxin-like 1 |
| chr11_+_107370375 | 0.19 |
ENSMUST00000106750.5
|
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr11_+_107370303 | 0.19 |
ENSMUST00000021063.13
ENSMUST00000106752.10 |
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr9_-_116004265 | 0.18 |
ENSMUST00000061101.12
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr3_-_144514386 | 0.18 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr12_-_46865709 | 0.17 |
ENSMUST00000021438.8
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr7_+_28580847 | 0.17 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr4_-_155947819 | 0.17 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr1_-_130866978 | 0.16 |
ENSMUST00000016668.13
|
Il19
|
interleukin 19 |
| chr13_-_50404344 | 0.16 |
ENSMUST00000099521.2
|
Gm906
|
predicted gene 906 |
| chr6_+_41192984 | 0.16 |
ENSMUST00000193997.6
ENSMUST00000103279.3 |
Trbv23
|
T cell receptor beta, variable V23 |
| chr16_-_50151350 | 0.16 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr5_+_20112500 | 0.16 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_+_40941688 | 0.16 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr3_-_89309944 | 0.15 |
ENSMUST00000057431.6
|
Lenep
|
lens epithelial protein |
| chr1_+_36730530 | 0.14 |
ENSMUST00000081180.7
ENSMUST00000193210.6 ENSMUST00000195151.6 |
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr13_+_112454981 | 0.14 |
ENSMUST00000223871.2
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr7_+_141048722 | 0.14 |
ENSMUST00000058746.7
|
Cd151
|
CD151 antigen |
| chr10_-_129948657 | 0.14 |
ENSMUST00000081469.2
|
Olfr823
|
olfactory receptor 823 |
| chr12_+_111132908 | 0.13 |
ENSMUST00000139162.8
ENSMUST00000060274.7 |
Traf3
|
TNF receptor-associated factor 3 |
| chr7_+_18915086 | 0.13 |
ENSMUST00000120595.8
ENSMUST00000048502.10 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr14_-_21798694 | 0.13 |
ENSMUST00000183943.2
|
Dusp13
|
dual specificity phosphatase 13 |
| chr9_+_107456086 | 0.12 |
ENSMUST00000149638.8
ENSMUST00000139274.8 ENSMUST00000130053.8 ENSMUST00000122985.8 ENSMUST00000127380.8 ENSMUST00000139581.2 |
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr7_+_18915136 | 0.12 |
ENSMUST00000144054.8
ENSMUST00000141718.8 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr11_+_69523754 | 0.12 |
ENSMUST00000018909.4
|
Fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr12_-_113760187 | 0.11 |
ENSMUST00000192911.2
ENSMUST00000103455.3 |
Ighv2-6-8
|
immunoglobulin heavy variable 2-6-8 |
| chr9_-_44437694 | 0.11 |
ENSMUST00000062215.8
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr5_-_73132045 | 0.11 |
ENSMUST00000043711.9
|
Gm10135
|
predicted gene 10135 |
| chr9_-_44437801 | 0.11 |
ENSMUST00000215661.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr2_+_90884612 | 0.11 |
ENSMUST00000185715.7
|
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
| chr2_+_90884349 | 0.10 |
ENSMUST00000067663.14
ENSMUST00000002171.14 ENSMUST00000111441.10 |
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
| chr4_-_138484889 | 0.10 |
ENSMUST00000030526.7
|
Pla2g2f
|
phospholipase A2, group IIF |
| chrX_+_132809189 | 0.09 |
ENSMUST00000113304.2
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chrX_+_163052367 | 0.09 |
ENSMUST00000145412.8
ENSMUST00000033749.9 |
Pir
|
pirin |
| chr6_-_68713748 | 0.09 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr3_+_130904000 | 0.09 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 0.7 | 3.6 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.7 | 4.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.7 | 5.5 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.7 | 7.4 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.6 | 5.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.4 | 1.2 | GO:1904432 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.4 | 1.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 1.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.3 | 5.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.3 | 1.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 1.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 2.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 1.2 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.5 | GO:1901874 | negative regulation of cellular respiration(GO:1901856) regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.2 | 0.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 1.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 2.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 1.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 2.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 2.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 7.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 1.2 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 1.8 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 1.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.3 | GO:0015881 | creatine transport(GO:0015881) |
| 0.1 | 0.5 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.9 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 0.3 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.9 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.6 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.4 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.6 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.3 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 1.9 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.1 | 2.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.1 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 | 0.7 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 3.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.5 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 1.5 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0060708 | histone H3-K27 acetylation(GO:0043974) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.4 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 2.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 2.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 2.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.5 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.2 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0061152 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) odontoblast differentiation(GO:0071895) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 1.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.3 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 1.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.4 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 3.4 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.4 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.7 | 2.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.6 | 5.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 1.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.2 | 0.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 2.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 0.5 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 1.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 4.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.3 | GO:1990590 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 4.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 3.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 1.9 | 5.7 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 1.8 | 7.4 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 1.8 | 5.5 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.9 | 5.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.7 | 3.6 | GO:0004854 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdopterin cofactor binding(GO:0043546) |
| 0.4 | 1.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 4.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 7.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 2.9 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 1.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 2.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.2 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 2.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.3 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 1.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 2.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 5.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0008127 | quercetin 2,3-dioxygenase activity(GO:0008127) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.9 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 5.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 6.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 6.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |