Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
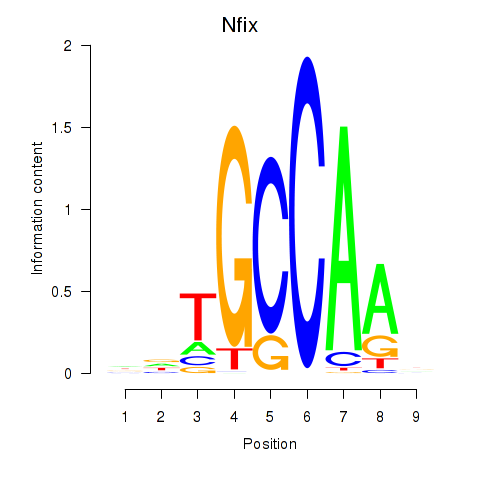
Results for Nfix
Z-value: 1.51
Transcription factors associated with Nfix
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfix
|
ENSMUSG00000001911.17 | Nfix |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfix | mm39_v1_chr8_-_85500998_85501108 | 0.57 | 2.7e-04 | Click! |
Activity profile of Nfix motif
Sorted Z-values of Nfix motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfix
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_97066937 | 8.15 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr7_+_140343652 | 5.83 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr7_+_130633776 | 5.59 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr7_-_126651847 | 5.26 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr5_+_115604321 | 5.09 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr8_-_93857899 | 4.87 |
ENSMUST00000034189.17
|
Ces1c
|
carboxylesterase 1C |
| chr6_-_141801897 | 4.63 |
ENSMUST00000165990.8
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr6_-_141801918 | 4.23 |
ENSMUST00000163678.2
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr19_-_43512929 | 2.68 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr19_-_46661321 | 2.49 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr9_-_103099262 | 2.40 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr5_-_77262968 | 2.39 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr19_-_46661501 | 2.33 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr17_-_74257164 | 2.26 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr19_-_40175709 | 2.20 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr5_+_90608751 | 2.20 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr6_-_78355834 | 2.14 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr10_+_57660948 | 2.13 |
ENSMUST00000020024.12
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr3_-_129126362 | 2.11 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr1_+_87998487 | 2.09 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr6_-_136852792 | 1.93 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr3_+_62245765 | 1.73 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr2_+_121978156 | 1.68 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chr1_+_133292898 | 1.65 |
ENSMUST00000129213.2
|
Etnk2
|
ethanolamine kinase 2 |
| chr1_-_162687369 | 1.64 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr5_-_87054796 | 1.60 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr11_+_101258368 | 1.52 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr5_-_137919873 | 1.51 |
ENSMUST00000031741.8
|
Cyp3a13
|
cytochrome P450, family 3, subfamily a, polypeptide 13 |
| chr17_-_35081129 | 1.48 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr17_-_35081456 | 1.47 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr7_-_48497771 | 1.46 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr11_-_100012384 | 1.39 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr1_+_133109059 | 1.34 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr10_+_116111441 | 1.28 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr10_-_89342493 | 1.23 |
ENSMUST00000058126.15
ENSMUST00000105296.9 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr10_-_127206300 | 1.22 |
ENSMUST00000026472.10
|
Inhbc
|
inhibin beta-C |
| chr11_+_68986043 | 1.21 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr11_+_115353290 | 1.19 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr7_-_25358406 | 1.18 |
ENSMUST00000071329.8
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr1_-_162726053 | 1.14 |
ENSMUST00000143123.3
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr11_+_67167950 | 1.14 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr9_+_106324952 | 1.10 |
ENSMUST00000215475.2
ENSMUST00000187106.7 ENSMUST00000190167.7 |
Abhd14b
|
abhydrolase domain containing 14b |
| chr2_+_133394079 | 1.08 |
ENSMUST00000028836.7
|
Bmp2
|
bone morphogenetic protein 2 |
| chr8_-_95405234 | 0.99 |
ENSMUST00000213043.2
|
Pllp
|
plasma membrane proteolipid |
| chr7_+_26895206 | 0.95 |
ENSMUST00000179391.8
ENSMUST00000108379.8 |
BC024978
|
cDNA sequence BC024978 |
| chr14_+_66074751 | 0.92 |
ENSMUST00000022614.7
|
Ccdc25
|
coiled-coil domain containing 25 |
| chr11_+_50917831 | 0.90 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr15_-_101621332 | 0.90 |
ENSMUST00000023709.7
|
Krt5
|
keratin 5 |
| chr6_+_78347844 | 0.89 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr11_+_67090878 | 0.87 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chrX_+_37405054 | 0.85 |
ENSMUST00000016471.9
ENSMUST00000115134.2 |
Atp1b4
|
ATPase, (Na+)/K+ transporting, beta 4 polypeptide |
| chr2_+_33106062 | 0.83 |
ENSMUST00000004208.7
|
Angptl2
|
angiopoietin-like 2 |
| chr3_+_122277372 | 0.82 |
ENSMUST00000197073.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr16_+_20492014 | 0.80 |
ENSMUST00000154950.8
ENSMUST00000115461.8 ENSMUST00000136713.5 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr8_-_23680954 | 0.77 |
ENSMUST00000209507.2
|
Gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr3_+_87855973 | 0.77 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr5_-_123887434 | 0.76 |
ENSMUST00000182955.8
ENSMUST00000182489.8 ENSMUST00000183147.9 ENSMUST00000050827.14 ENSMUST00000057795.12 ENSMUST00000111515.8 ENSMUST00000182309.8 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chrX_-_47297436 | 0.75 |
ENSMUST00000037960.11
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr15_-_101833160 | 0.73 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr1_+_152275575 | 0.73 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr16_+_42727926 | 0.72 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_172047282 | 0.72 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chr4_+_105014536 | 0.72 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr2_+_34661982 | 0.71 |
ENSMUST00000028222.13
ENSMUST00000100171.3 |
Hspa5
|
heat shock protein 5 |
| chr2_+_33106121 | 0.70 |
ENSMUST00000193373.3
|
Angptl2
|
angiopoietin-like 2 |
| chr6_+_34723304 | 0.70 |
ENSMUST00000142716.3
|
Cald1
|
caldesmon 1 |
| chrX_-_8059597 | 0.69 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr1_-_155293141 | 0.68 |
ENSMUST00000111775.8
ENSMUST00000111774.2 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr10_+_57661010 | 0.68 |
ENSMUST00000165013.2
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr16_+_22676589 | 0.68 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr3_+_89987749 | 0.67 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chrX_-_105647282 | 0.65 |
ENSMUST00000113480.2
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr6_-_33037107 | 0.65 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr3_+_63203235 | 0.65 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr4_+_102427247 | 0.64 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chrX_-_111315519 | 0.64 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr2_-_69172944 | 0.64 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr4_-_123466853 | 0.61 |
ENSMUST00000238555.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr10_+_4561974 | 0.61 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr8_+_91554933 | 0.61 |
ENSMUST00000048665.8
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr3_-_144738526 | 0.60 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr2_-_113883285 | 0.60 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chrX_-_50294652 | 0.60 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr6_-_33037191 | 0.60 |
ENSMUST00000066379.11
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr6_-_5256282 | 0.59 |
ENSMUST00000031773.9
|
Pon3
|
paraoxonase 3 |
| chr13_+_120151915 | 0.59 |
ENSMUST00000225543.2
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr9_+_43978369 | 0.58 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr7_+_126396779 | 0.58 |
ENSMUST00000205324.2
|
Tlcd3b
|
TLC domain containing 3B |
| chr19_+_8828132 | 0.58 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr4_-_141351110 | 0.57 |
ENSMUST00000038661.8
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr3_+_99203818 | 0.55 |
ENSMUST00000150756.3
|
Tbx15
|
T-box 15 |
| chr16_+_51851917 | 0.55 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr12_+_36042899 | 0.55 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr2_-_5680801 | 0.55 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr13_+_25127127 | 0.54 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr1_+_164624200 | 0.54 |
ENSMUST00000027861.6
|
Dpt
|
dermatopontin |
| chr19_-_11806388 | 0.53 |
ENSMUST00000061235.3
|
Olfr1417
|
olfactory receptor 1417 |
| chr14_-_31503869 | 0.53 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr19_+_56276375 | 0.53 |
ENSMUST00000166049.8
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr5_-_151351628 | 0.53 |
ENSMUST00000202365.2
ENSMUST00000186838.2 |
Gm42906
D730045B01Rik
|
predicted gene 42906 RIKEN cDNA D730045B01 gene |
| chrX_+_162691978 | 0.52 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chrX_-_99456185 | 0.51 |
ENSMUST00000033567.15
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr7_-_5128936 | 0.51 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chrX_-_47297746 | 0.51 |
ENSMUST00000088935.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr16_+_51851588 | 0.50 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr11_+_98274637 | 0.49 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap |
| chr11_+_77353431 | 0.49 |
ENSMUST00000130255.2
|
Coro6
|
coronin 6 |
| chr6_+_15727798 | 0.49 |
ENSMUST00000128849.3
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr4_-_129121676 | 0.49 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr5_-_31250817 | 0.49 |
ENSMUST00000031037.14
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr7_+_114344920 | 0.49 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr4_+_95467701 | 0.48 |
ENSMUST00000150830.2
ENSMUST00000134012.9 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chrX_+_100420873 | 0.47 |
ENSMUST00000052130.14
|
Gjb1
|
gap junction protein, beta 1 |
| chr14_-_36628263 | 0.47 |
ENSMUST00000183007.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_-_162687488 | 0.47 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr11_-_83959380 | 0.46 |
ENSMUST00000164891.8
|
Dusp14
|
dual specificity phosphatase 14 |
| chr6_-_115228800 | 0.45 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr19_+_56276343 | 0.44 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr10_-_125164826 | 0.43 |
ENSMUST00000211781.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr1_-_57008986 | 0.43 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr7_+_140425460 | 0.43 |
ENSMUST00000035300.7
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr12_+_84687721 | 0.43 |
ENSMUST00000221915.2
ENSMUST00000166772.9 |
Vrtn
|
vertebrae development associated |
| chr17_+_45817750 | 0.43 |
ENSMUST00000024733.9
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr5_+_92535705 | 0.43 |
ENSMUST00000138687.2
ENSMUST00000124509.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr14_+_65903840 | 0.43 |
ENSMUST00000022610.15
|
Scara5
|
scavenger receptor class A, member 5 |
| chr12_-_84408576 | 0.43 |
ENSMUST00000021659.2
ENSMUST00000065536.9 |
Fam161b
|
family with sequence similarity 161, member B |
| chr9_+_89791943 | 0.42 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr10_-_12743915 | 0.42 |
ENSMUST00000219584.2
|
Utrn
|
utrophin |
| chr13_+_93440265 | 0.42 |
ENSMUST00000109494.8
|
Homer1
|
homer scaffolding protein 1 |
| chr10_-_75658355 | 0.42 |
ENSMUST00000160211.2
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr10_-_12744025 | 0.41 |
ENSMUST00000219660.2
|
Utrn
|
utrophin |
| chr13_+_42862957 | 0.39 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr5_+_149335214 | 0.39 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr3_-_121076745 | 0.39 |
ENSMUST00000135818.8
ENSMUST00000137234.2 |
Tlcd4
|
TLC domain containing 4 |
| chr15_-_78090591 | 0.39 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin |
| chr10_-_89093441 | 0.38 |
ENSMUST00000182341.8
ENSMUST00000182613.8 |
Ano4
|
anoctamin 4 |
| chr19_-_34856853 | 0.38 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr18_-_23171713 | 0.38 |
ENSMUST00000081423.13
|
Nol4
|
nucleolar protein 4 |
| chr10_-_105410280 | 0.38 |
ENSMUST00000061506.9
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr16_+_41353212 | 0.38 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr2_-_142743354 | 0.38 |
ENSMUST00000211861.2
|
Kif16b
|
kinesin family member 16B |
| chrX_-_111316476 | 0.37 |
ENSMUST00000026601.3
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr5_+_92535532 | 0.37 |
ENSMUST00000145072.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr11_+_67689094 | 0.37 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chrX_+_162692126 | 0.37 |
ENSMUST00000033734.14
ENSMUST00000112294.9 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_-_22930188 | 0.37 |
ENSMUST00000114544.10
ENSMUST00000139038.8 ENSMUST00000126112.8 ENSMUST00000178908.2 ENSMUST00000078977.14 ENSMUST00000140164.8 ENSMUST00000149719.8 |
Abi1
|
abl interactor 1 |
| chr7_+_27879650 | 0.37 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr11_-_78313043 | 0.37 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr6_+_78347636 | 0.37 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr2_-_147028309 | 0.36 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr8_-_37200051 | 0.36 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr9_+_21095399 | 0.36 |
ENSMUST00000115458.9
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr15_+_10952418 | 0.36 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr3_-_7678785 | 0.35 |
ENSMUST00000194279.6
|
Il7
|
interleukin 7 |
| chr4_-_129155185 | 0.35 |
ENSMUST00000145261.8
|
C77080
|
expressed sequence C77080 |
| chr8_+_91555449 | 0.35 |
ENSMUST00000109614.9
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr17_-_15163362 | 0.35 |
ENSMUST00000238668.2
ENSMUST00000228330.2 |
Wdr27
|
WD repeat domain 27 |
| chr8_+_31581635 | 0.35 |
ENSMUST00000161713.2
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr1_+_93271340 | 0.34 |
ENSMUST00000185498.7
|
Ppp1r7
|
protein phosphatase 1, regulatory subunit 7 |
| chr2_+_20742115 | 0.34 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr16_-_3726503 | 0.34 |
ENSMUST00000115859.8
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr11_+_115802828 | 0.33 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr9_-_87613301 | 0.33 |
ENSMUST00000034991.8
|
Tbx18
|
T-box18 |
| chr2_-_160155536 | 0.33 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr5_-_104125226 | 0.33 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_+_24618380 | 0.33 |
ENSMUST00000049346.10
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chr12_-_45120895 | 0.30 |
ENSMUST00000120531.8
ENSMUST00000143376.8 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr5_-_147662798 | 0.30 |
ENSMUST00000110529.6
ENSMUST00000031653.12 |
Flt1
|
FMS-like tyrosine kinase 1 |
| chr6_-_5256225 | 0.30 |
ENSMUST00000125686.8
|
Pon3
|
paraoxonase 3 |
| chr12_-_70394074 | 0.30 |
ENSMUST00000223160.2
ENSMUST00000222316.2 ENSMUST00000167755.3 ENSMUST00000110520.10 ENSMUST00000110522.10 ENSMUST00000221041.2 ENSMUST00000222603.3 |
Trim9
|
tripartite motif-containing 9 |
| chr19_-_11833365 | 0.30 |
ENSMUST00000079875.4
|
Olfr1418
|
olfactory receptor 1418 |
| chr7_+_133239272 | 0.30 |
ENSMUST00000051169.13
|
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr12_-_55539372 | 0.30 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr15_-_64794139 | 0.30 |
ENSMUST00000023007.7
ENSMUST00000228014.2 |
Adcy8
|
adenylate cyclase 8 |
| chr13_+_49761506 | 0.29 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr7_+_133239414 | 0.29 |
ENSMUST00000128901.9
|
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr11_+_49500090 | 0.29 |
ENSMUST00000020617.3
|
Flt4
|
FMS-like tyrosine kinase 4 |
| chr6_+_113460258 | 0.28 |
ENSMUST00000032422.6
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr11_+_69047815 | 0.28 |
ENSMUST00000036424.3
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type |
| chr5_-_104125270 | 0.28 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr10_+_21869776 | 0.28 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_+_141276575 | 0.28 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr11_-_43727071 | 0.28 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chrX_+_113384297 | 0.28 |
ENSMUST00000133447.2
|
Klhl4
|
kelch-like 4 |
| chr3_-_89186940 | 0.27 |
ENSMUST00000118860.2
ENSMUST00000029566.9 |
Efna1
|
ephrin A1 |
| chr1_-_86353932 | 0.27 |
ENSMUST00000212541.2
|
Nmur1
|
neuromedin U receptor 1 |
| chr3_-_7678796 | 0.27 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr11_-_114686712 | 0.27 |
ENSMUST00000000206.4
|
Btbd17
|
BTB (POZ) domain containing 17 |
| chr15_-_98769056 | 0.27 |
ENSMUST00000178486.9
ENSMUST00000023741.16 |
Kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr6_+_83055581 | 0.27 |
ENSMUST00000177177.8
ENSMUST00000176089.2 |
Pcgf1
|
polycomb group ring finger 1 |
| chrX_-_50294867 | 0.27 |
ENSMUST00000114876.9
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr1_+_169757498 | 0.26 |
ENSMUST00000175731.3
|
Ccdc190
|
coiled-coil domain containing 190 |
| chr9_-_123546605 | 0.26 |
ENSMUST00000163207.2
|
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr6_-_125290809 | 0.26 |
ENSMUST00000032489.8
|
Ltbr
|
lymphotoxin B receptor |
| chr10_-_125164399 | 0.26 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr17_-_14279793 | 0.26 |
ENSMUST00000186636.3
|
Gm7358
|
predicted gene 7358 |
| chr4_-_42661893 | 0.26 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr5_-_104125192 | 0.25 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr18_+_34380738 | 0.25 |
ENSMUST00000066133.7
|
Apc
|
APC, WNT signaling pathway regulator |
| chr11_-_84771021 | 0.25 |
ENSMUST00000067058.3
ENSMUST00000108080.3 |
Pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.9 | 2.7 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.7 | 2.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.6 | 1.7 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.5 | 1.5 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.5 | 2.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.3 | 9.1 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.3 | 1.2 | GO:0034971 | histone H3-R17 methylation(GO:0034971) bile acid signaling pathway(GO:0038183) |
| 0.3 | 2.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.3 | 2.4 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 1.1 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.2 | 4.8 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.2 | 0.7 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 0.9 | GO:0046226 | coumarin metabolic process(GO:0009804) coumarin catabolic process(GO:0046226) |
| 0.2 | 2.4 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 5.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 2.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 8.6 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.2 | 1.0 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 1.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 3.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 9.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.8 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.2 | 0.6 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.3 | GO:2000729 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 1.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.7 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.1 | 0.9 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.6 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 1.5 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 1.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 2.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.6 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 1.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 1.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.7 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.7 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.3 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.1 | 0.5 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 1.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 2.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.3 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.5 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.3 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.4 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 0.2 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.1 | 1.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.5 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 0.4 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 1.1 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 1.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.3 | GO:0050968 | sensory perception of sour taste(GO:0050915) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.2 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.4 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.0 | 0.3 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.2 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.5 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.2 | GO:0060578 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.6 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.2 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.6 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.5 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.6 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 1.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.3 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.7 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.6 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.0 | 0.5 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.7 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.9 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.9 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.5 | GO:0060487 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 4.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 0.9 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.2 | 0.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 1.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 1.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 2.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.4 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.1 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 2.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 8.1 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 4.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.3 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 5.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 22.4 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.6 | 5.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 2.3 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.4 | 9.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.4 | 1.6 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.4 | 1.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.4 | 1.5 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 2.4 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 3.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 9.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 1.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 4.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 0.9 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.2 | 1.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.6 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 0.5 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.5 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 2.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.5 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.1 | 0.7 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 3.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 4.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 4.3 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 6.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 6.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.6 | 1.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 4.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 5.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 2.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |