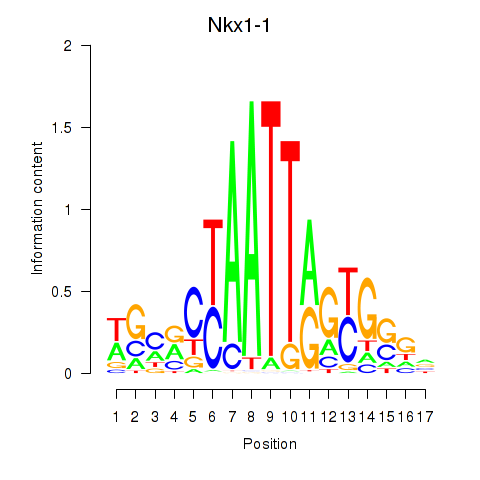
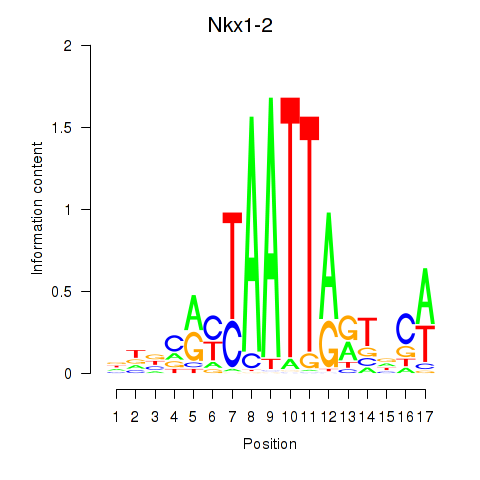
|
chr4_-_131802561
Show fit
|
1.74 |
ENSMUST00000105970.8
ENSMUST00000105975.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr6_+_29694181
Show fit
|
1.61 |
ENSMUST00000046750.14
ENSMUST00000115250.4
|
Tspan33
|
tetraspanin 33
|
|
chr4_-_131802606
Show fit
|
1.53 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr11_-_86999481
Show fit
|
1.47 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11
|
|
chr9_-_20871081
Show fit
|
1.14 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1
|
|
chr14_+_46997984
Show fit
|
1.07 |
ENSMUST00000067426.6
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3
|
|
chr1_-_173161069
Show fit
|
1.03 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group)
|
|
chr7_-_44578834
Show fit
|
1.02 |
ENSMUST00000107857.11
ENSMUST00000167930.8
ENSMUST00000085399.13
ENSMUST00000166972.9
|
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit
|
|
chr11_+_87000032
Show fit
|
0.94 |
ENSMUST00000020794.6
|
Ska2
|
spindle and kinetochore associated complex subunit 2
|
|
chr11_+_43572825
Show fit
|
0.76 |
ENSMUST00000061070.6
ENSMUST00000094294.5
|
Pwwp2a
|
PWWP domain containing 2A
|
|
chr11_+_99748741
Show fit
|
0.72 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568
|
|
chr5_+_30824121
Show fit
|
0.69 |
ENSMUST00000144742.6
ENSMUST00000149759.2
ENSMUST00000199320.5
|
Cenpa
|
centromere protein A
|
|
chr14_+_46998004
Show fit
|
0.66 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3
|
|
chr11_+_121128042
Show fit
|
0.65 |
ENSMUST00000103015.4
|
Narf
|
nuclear prelamin A recognition factor
|
|
chrX_+_9751861
Show fit
|
0.62 |
ENSMUST00000067529.9
ENSMUST00000086165.4
|
Sytl5
|
synaptotagmin-like 5
|
|
chr1_+_82817794
Show fit
|
0.57 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1
|
|
chr4_-_140501507
Show fit
|
0.52 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV
|
|
chr2_+_36120438
Show fit
|
0.49 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1
|
|
chr3_-_90297187
Show fit
|
0.46 |
ENSMUST00000029541.12
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3
|
|
chr14_+_65612788
Show fit
|
0.45 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395
|
|
chr1_-_91386976
Show fit
|
0.44 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2
|
|
chr19_+_5540483
Show fit
|
0.44 |
ENSMUST00000209469.2
ENSMUST00000116560.3
|
Cfl1
|
cofilin 1, non-muscle
|
|
chr6_-_128339775
Show fit
|
0.44 |
ENSMUST00000112152.8
ENSMUST00000057421.15
ENSMUST00000112151.2
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1
|
|
chr5_-_122959361
Show fit
|
0.43 |
ENSMUST00000086216.9
|
Anapc5
|
anaphase-promoting complex subunit 5
|
|
chr7_+_29683373
Show fit
|
0.42 |
ENSMUST00000148442.8
|
Zfp568
|
zinc finger protein 568
|
|
chr12_-_40087393
Show fit
|
0.42 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A
|
|
chr4_-_88595161
Show fit
|
0.42 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16
|
|
chr9_-_44024767
Show fit
|
0.40 |
ENSMUST00000216511.2
ENSMUST00000056328.6
ENSMUST00000185479.2
|
Rnf26
Gm49380
|
ring finger protein 26
predicted gene, 49380
|
|
chr19_-_40371016
Show fit
|
0.37 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr14_-_52341426
Show fit
|
0.36 |
ENSMUST00000227536.2
ENSMUST00000227195.2
ENSMUST00000228815.2
ENSMUST00000228198.2
ENSMUST00000227458.2
ENSMUST00000228232.2
ENSMUST00000227242.2
ENSMUST00000228748.2
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C
|
|
chr5_-_137101108
Show fit
|
0.36 |
ENSMUST00000077523.4
ENSMUST00000041388.11
|
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1
|
|
chr10_+_128173603
Show fit
|
0.32 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase
|
|
chr19_+_5540591
Show fit
|
0.32 |
ENSMUST00000237122.2
|
Cfl1
|
cofilin 1, non-muscle
|
|
chr14_-_52341472
Show fit
|
0.31 |
ENSMUST00000111610.12
ENSMUST00000164655.2
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C
|
|
chr9_-_66033841
Show fit
|
0.30 |
ENSMUST00000137542.2
|
Snx1
|
sorting nexin 1
|
|
chr3_-_89009153
Show fit
|
0.28 |
ENSMUST00000199668.3
ENSMUST00000196709.5
|
Fdps
|
farnesyl diphosphate synthetase
|
|
chr18_+_4993795
Show fit
|
0.27 |
ENSMUST00000153016.8
|
Svil
|
supervillin
|
|
chr5_+_143846782
Show fit
|
0.27 |
ENSMUST00000148011.8
ENSMUST00000110709.7
|
Pms2
|
PMS1 homolog2, mismatch repair system component
|
|
chr19_+_41921903
Show fit
|
0.27 |
ENSMUST00000224258.2
ENSMUST00000026154.9
ENSMUST00000224896.2
|
Zdhhc16
|
zinc finger, DHHC domain containing 16
|
|
chr7_+_101545547
Show fit
|
0.27 |
ENSMUST00000035395.14
ENSMUST00000106973.8
ENSMUST00000144207.9
|
Anapc15
|
anaphase promoting complex C subunit 15
|
|
chr4_-_43710231
Show fit
|
0.27 |
ENSMUST00000217544.2
ENSMUST00000107862.3
|
Olfr71
|
olfactory receptor 71
|
|
chr17_-_57338468
Show fit
|
0.27 |
ENSMUST00000007814.10
ENSMUST00000233480.2
|
Khsrp
|
KH-type splicing regulatory protein
|
|
chr11_+_104468107
Show fit
|
0.26 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4
|
|
chr7_-_110673269
Show fit
|
0.26 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2
|
|
chr9_+_50686647
Show fit
|
0.26 |
ENSMUST00000159576.2
|
Alg9
|
asparagine-linked glycosylation 9 (alpha 1,2 mannosyltransferase)
|
|
chr12_+_17778198
Show fit
|
0.25 |
ENSMUST00000222944.2
|
Hpcal1
|
hippocalcin-like 1
|
|
chr3_-_89009214
Show fit
|
0.24 |
ENSMUST00000081848.13
|
Fdps
|
farnesyl diphosphate synthetase
|
|
chr7_-_100613579
Show fit
|
0.24 |
ENSMUST00000060174.6
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr19_+_60744385
Show fit
|
0.23 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1
|
|
chr2_+_154390808
Show fit
|
0.23 |
ENSMUST00000045116.11
ENSMUST00000109709.4
|
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene
|
|
chr7_-_26895141
Show fit
|
0.23 |
ENSMUST00000163311.9
ENSMUST00000126211.2
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A
|
|
chr2_+_85809620
Show fit
|
0.22 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030
|
|
chr15_+_97990431
Show fit
|
0.21 |
ENSMUST00000229280.2
ENSMUST00000163507.8
ENSMUST00000230445.2
|
Pfkm
|
phosphofructokinase, muscle
|
|
chr7_+_101546059
Show fit
|
0.21 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15
|
|
chr11_-_99121822
Show fit
|
0.21 |
ENSMUST00000103133.4
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1
|
|
chr6_-_128339458
Show fit
|
0.20 |
ENSMUST00000155573.3
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1
|
|
chr7_+_103628383
Show fit
|
0.20 |
ENSMUST00000098185.2
|
Olfr635
|
olfactory receptor 635
|
|
chr2_-_168607166
Show fit
|
0.19 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4
|
|
chr15_-_77840856
Show fit
|
0.19 |
ENSMUST00000117725.2
ENSMUST00000016696.13
|
Foxred2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr5_+_138115165
Show fit
|
0.19 |
ENSMUST00000062350.15
ENSMUST00000110961.9
ENSMUST00000080732.10
ENSMUST00000110960.9
ENSMUST00000142185.8
ENSMUST00000136425.2
ENSMUST00000110959.2
|
Zscan21
|
zinc finger and SCAN domain containing 21
|
|
chr19_-_29344694
Show fit
|
0.18 |
ENSMUST00000138051.2
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein
|
|
chr11_+_104467791
Show fit
|
0.18 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4
|
|
chr3_+_94320548
Show fit
|
0.17 |
ENSMUST00000166032.8
ENSMUST00000200486.5
ENSMUST00000196386.5
ENSMUST00000045245.10
ENSMUST00000197901.5
ENSMUST00000198041.2
|
Tdrkh
Gm42463
|
tudor and KH domain containing protein
predicted gene 42463
|
|
chr19_+_29344846
Show fit
|
0.17 |
ENSMUST00000016640.8
|
Cd274
|
CD274 antigen
|
|
chr7_+_128346655
Show fit
|
0.16 |
ENSMUST00000042942.10
|
Sec23ip
|
Sec23 interacting protein
|
|
chr17_+_7437500
Show fit
|
0.16 |
ENSMUST00000024575.8
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2
|
|
chr2_-_45002902
Show fit
|
0.16 |
ENSMUST00000076836.13
ENSMUST00000176732.8
ENSMUST00000200844.4
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_111843053
Show fit
|
0.15 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310
|
|
chr9_-_85631361
Show fit
|
0.15 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase
|
|
chr1_-_131441962
Show fit
|
0.15 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2
|
|
chr11_-_99134885
Show fit
|
0.15 |
ENSMUST00000103132.10
ENSMUST00000038214.7
|
Krt222
|
keratin 222
|
|
chr10_+_94411119
Show fit
|
0.15 |
ENSMUST00000121471.8
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr6_-_68887922
Show fit
|
0.14 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86
|
|
chr5_+_134961246
Show fit
|
0.14 |
ENSMUST00000111218.8
ENSMUST00000136246.5
ENSMUST00000201847.3
|
Mettl27
|
methyltransferase like 27
|
|
chr19_+_8779903
Show fit
|
0.14 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3
|
|
chr18_-_24736848
Show fit
|
0.14 |
ENSMUST00000070726.10
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6
|
|
chr13_+_94219934
Show fit
|
0.14 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2
|
|
chr1_+_83022653
Show fit
|
0.14 |
ENSMUST00000222567.2
|
Gm47969
|
predicted gene, 47969
|
|
chr2_+_110427643
Show fit
|
0.14 |
ENSMUST00000045972.13
ENSMUST00000111026.3
|
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr10_+_80973787
Show fit
|
0.13 |
ENSMUST00000117956.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a
|
|
chr11_-_99742434
Show fit
|
0.13 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16
|
|
chr6_-_57340712
Show fit
|
0.12 |
ENSMUST00000228156.2
ENSMUST00000227186.2
ENSMUST00000228294.2
ENSMUST00000228342.2
|
Vmn1r17
|
vomeronasal 1 receptor 17
|
|
chr15_+_16728842
Show fit
|
0.12 |
ENSMUST00000228307.2
|
Cdh9
|
cadherin 9
|
|
chr7_-_30251680
Show fit
|
0.12 |
ENSMUST00000215288.2
ENSMUST00000108165.8
ENSMUST00000153594.3
|
Proser3
|
proline and serine rich 3
|
|
chr3_+_75982890
Show fit
|
0.12 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5
|
|
chr6_-_58452315
Show fit
|
0.12 |
ENSMUST00000177318.3
ENSMUST00000176147.9
ENSMUST00000176023.3
ENSMUST00000228586.2
|
Vmn1r31
|
vomeronasal 1 receptor 31
|
|
chr18_-_24736521
Show fit
|
0.11 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6
|
|
chr19_-_24178000
Show fit
|
0.10 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2
|
|
chr5_-_143846600
Show fit
|
0.10 |
ENSMUST00000031613.11
ENSMUST00000100483.3
|
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2
|
|
chr1_-_165462020
Show fit
|
0.10 |
ENSMUST00000194437.6
ENSMUST00000068705.13
ENSMUST00000111435.9
ENSMUST00000193023.2
|
Mpzl1
|
myelin protein zero-like 1
|
|
chr1_+_173093568
Show fit
|
0.10 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418
|
|
chr4_-_141327146
Show fit
|
0.08 |
ENSMUST00000141518.8
ENSMUST00000127455.8
ENSMUST00000105784.8
|
Fblim1
|
filamin binding LIM protein 1
|
|
chr8_+_34089597
Show fit
|
0.07 |
ENSMUST00000009774.11
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform
|
|
chr14_+_13803477
Show fit
|
0.07 |
ENSMUST00000102996.4
|
Cfap20dc
|
CFAP20 domain containing
|
|
chr2_-_165230165
Show fit
|
0.07 |
ENSMUST00000103084.4
|
Zfp334
|
zinc finger protein 334
|
|
chr10_+_97442727
Show fit
|
0.06 |
ENSMUST00000105286.4
|
Kera
|
keratocan
|
|
chr6_-_23650205
Show fit
|
0.06 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133
|
|
chr1_+_172383499
Show fit
|
0.06 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8
|
|
chr14_+_25980463
Show fit
|
0.06 |
ENSMUST00000173155.8
|
Duxbl1
|
double homeobox B-like 1
|
|
chr16_+_35861554
Show fit
|
0.05 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B
|
|
chrX_+_56257374
Show fit
|
0.05 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand
|
|
chr15_+_41694317
Show fit
|
0.05 |
ENSMUST00000166917.3
ENSMUST00000230127.2
ENSMUST00000230131.2
|
Oxr1
|
oxidation resistance 1
|
|
chr19_+_12364643
Show fit
|
0.05 |
ENSMUST00000217062.3
ENSMUST00000216145.2
ENSMUST00000213657.2
|
Olfr1440
|
olfactory receptor 1440
|
|
chr2_-_157188568
Show fit
|
0.05 |
ENSMUST00000029172.2
|
Ghrh
|
growth hormone releasing hormone
|
|
chr15_+_81548090
Show fit
|
0.05 |
ENSMUST00000023029.15
ENSMUST00000174229.8
ENSMUST00000172748.8
|
L3mbtl2
|
L3MBTL2 polycomb repressive complex 1 subunit
|
|
chr17_-_37430949
Show fit
|
0.05 |
ENSMUST00000214994.2
ENSMUST00000216341.2
|
Olfr92
|
olfactory receptor 92
|
|
chr7_-_11414074
Show fit
|
0.05 |
ENSMUST00000227010.2
ENSMUST00000209638.3
|
Vmn1r72
|
vomeronasal 1 receptor 72
|
|
chr6_-_128339654
Show fit
|
0.04 |
ENSMUST00000203719.2
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1
|
|
chr4_-_119151717
Show fit
|
0.04 |
ENSMUST00000079644.13
|
Ybx1
|
Y box protein 1
|
|
chr2_-_89855921
Show fit
|
0.04 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264
|
|
chrX_-_73129195
Show fit
|
0.04 |
ENSMUST00000140399.3
ENSMUST00000123362.8
ENSMUST00000100750.10
ENSMUST00000033770.13
|
Mecp2
|
methyl CpG binding protein 2
|
|
chr1_+_190660689
Show fit
|
0.04 |
ENSMUST00000066632.14
ENSMUST00000110899.7
|
Angel2
|
angel homolog 2
|
|
chr19_-_12313274
Show fit
|
0.04 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1
|
|
chr14_-_52277310
Show fit
|
0.04 |
ENSMUST00000216907.2
ENSMUST00000214071.2
ENSMUST00000216188.2
|
Olfr221
|
olfactory receptor 221
|
|
chr7_+_103652466
Show fit
|
0.03 |
ENSMUST00000209757.3
|
Olfr638
|
olfactory receptor 638
|
|
chr12_+_108145802
Show fit
|
0.03 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K
|
|
chr12_+_84463970
Show fit
|
0.03 |
ENSMUST00000183146.2
|
Rnf113a2
|
ring finger protein 113A2
|
|
chr16_-_29363671
Show fit
|
0.02 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4
|
|
chr5_+_134961205
Show fit
|
0.02 |
ENSMUST00000047196.14
ENSMUST00000111221.9
ENSMUST00000111219.8
ENSMUST00000068617.12
|
Mettl27
|
methyltransferase like 27
|
|
chrX_+_159551009
Show fit
|
0.02 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human)
|
|
chr10_+_129539079
Show fit
|
0.02 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804
|
|
chr12_+_52746158
Show fit
|
0.02 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6
|
|
chr11_-_22236795
Show fit
|
0.01 |
ENSMUST00000180360.8
ENSMUST00000109563.9
|
Ehbp1
|
EH domain binding protein 1
|
|
chr8_+_22329942
Show fit
|
0.01 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2
|
|
chr6_-_129752812
Show fit
|
0.01 |
ENSMUST00000095409.3
|
Klrh1
|
killer cell lectin-like receptor subfamily H, member 1
|
|
chrX_-_158921370
Show fit
|
0.01 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1
|
|
chr2_+_85804239
Show fit
|
0.01 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029
|
|
chr2_-_102230602
Show fit
|
0.00 |
ENSMUST00000152929.2
|
Trim44
|
tripartite motif-containing 44
|