Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
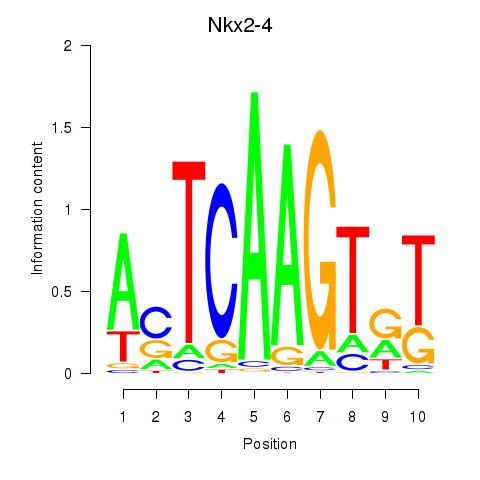
Results for Nkx2-4
Z-value: 0.45
Transcription factors associated with Nkx2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-4
|
ENSMUSG00000054160.3 | Nkx2-4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-4 | mm39_v1_chr2_-_146927365_146927406 | -0.02 | 9.3e-01 | Click! |
Activity profile of Nkx2-4 motif
Sorted Z-values of Nkx2-4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_26821266 | 1.98 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr6_+_41331039 | 1.01 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr19_-_39875192 | 0.90 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr19_-_39801188 | 0.69 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr19_-_39637489 | 0.60 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr8_-_110305672 | 0.41 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr17_+_37253802 | 0.27 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr4_-_40279382 | 0.25 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr5_+_32293145 | 0.23 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
| chr6_+_41098273 | 0.23 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr3_-_10366229 | 0.21 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr7_-_44320244 | 0.21 |
ENSMUST00000048102.15
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr15_+_88703786 | 0.20 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr1_-_175319842 | 0.19 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr3_-_102843406 | 0.19 |
ENSMUST00000199930.2
ENSMUST00000029448.11 ENSMUST00000196988.5 |
Sycp1
|
synaptonemal complex protein 1 |
| chr9_-_31824758 | 0.18 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chrX_+_100427331 | 0.18 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr2_-_63014622 | 0.17 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_-_157408239 | 0.17 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr18_-_3281089 | 0.16 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr2_-_147028309 | 0.14 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr4_+_48663504 | 0.14 |
ENSMUST00000030033.5
|
Cavin4
|
caveolae associated 4 |
| chr19_-_45731290 | 0.14 |
ENSMUST00000111927.8
|
Fgf8
|
fibroblast growth factor 8 |
| chr4_+_99544536 | 0.14 |
ENSMUST00000087285.5
|
Foxd3
|
forkhead box D3 |
| chr13_+_63387870 | 0.13 |
ENSMUST00000159152.3
ENSMUST00000221820.2 |
Aopep
|
aminopeptidase O |
| chr19_-_39729431 | 0.13 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr13_+_63387827 | 0.13 |
ENSMUST00000222929.2
|
Aopep
|
aminopeptidase O |
| chrX_+_102674181 | 0.12 |
ENSMUST00000033692.9
|
Zcchc13
|
zinc finger, CCHC domain containing 13 |
| chr1_+_85177316 | 0.12 |
ENSMUST00000161424.5
ENSMUST00000113402.4 |
Gm7609
|
predicted pseudogene 7609 |
| chr8_+_46081213 | 0.12 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr5_+_14564932 | 0.11 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chrX_+_106132840 | 0.11 |
ENSMUST00000118666.8
ENSMUST00000053375.4 |
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr8_-_40761372 | 0.11 |
ENSMUST00000118639.2
|
Fgf20
|
fibroblast growth factor 20 |
| chr12_+_105651643 | 0.11 |
ENSMUST00000051934.7
|
Gskip
|
GSK3B interacting protein |
| chr18_-_3281727 | 0.11 |
ENSMUST00000154705.8
ENSMUST00000151084.8 |
Crem
|
cAMP responsive element modulator |
| chr11_-_102470287 | 0.10 |
ENSMUST00000107081.8
|
Gm11627
|
predicted gene 11627 |
| chrX_-_139857424 | 0.10 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr3_-_92496293 | 0.10 |
ENSMUST00000098888.7
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chrX_-_142716085 | 0.10 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr16_-_92622659 | 0.10 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr19_-_45731312 | 0.09 |
ENSMUST00000026241.12
ENSMUST00000026240.14 ENSMUST00000111928.8 |
Fgf8
|
fibroblast growth factor 8 |
| chr11_-_83320281 | 0.09 |
ENSMUST00000052521.9
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr2_+_61634797 | 0.09 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr1_+_85454323 | 0.09 |
ENSMUST00000239236.2
|
Gm7592
|
predicted gene 7592 |
| chr2_-_35995283 | 0.09 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr10_-_62258195 | 0.08 |
ENSMUST00000020277.9
|
Hkdc1
|
hexokinase domain containing 1 |
| chr6_+_121320339 | 0.08 |
ENSMUST00000168295.2
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr3_-_129548954 | 0.08 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr11_+_53611611 | 0.08 |
ENSMUST00000048605.3
|
Il5
|
interleukin 5 |
| chr18_-_3281752 | 0.08 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chrX_-_35755483 | 0.08 |
ENSMUST00000169499.2
|
Gm14569
|
predicted gene 14569 |
| chr15_+_81350842 | 0.08 |
ENSMUST00000230062.2
|
Rbx1
|
ring-box 1 |
| chr14_+_55909816 | 0.08 |
ENSMUST00000227178.2
ENSMUST00000227914.2 |
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr17_+_37253916 | 0.08 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr14_+_26414422 | 0.08 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr14_+_55909692 | 0.08 |
ENSMUST00000002397.7
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr18_+_37085673 | 0.07 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr10_+_60236532 | 0.07 |
ENSMUST00000218360.2
|
Gm17455
|
predicted gene, 17455 |
| chrX_+_36572501 | 0.07 |
ENSMUST00000179915.4
|
Rhox4a2
|
reproductive homeobox 4A2 |
| chr2_+_153716958 | 0.07 |
ENSMUST00000028983.3
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr11_+_110858882 | 0.07 |
ENSMUST00000125692.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_+_16175998 | 0.07 |
ENSMUST00000027053.8
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr8_+_46944000 | 0.07 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr11_-_59029996 | 0.06 |
ENSMUST00000219084.3
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr15_+_21111428 | 0.06 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr1_-_63153675 | 0.06 |
ENSMUST00000097718.9
|
Ino80d
|
INO80 complex subunit D |
| chr1_+_59669482 | 0.06 |
ENSMUST00000190490.2
|
Gm973
|
predicted gene 973 |
| chr11_-_53750016 | 0.06 |
ENSMUST00000117316.8
ENSMUST00000120776.8 ENSMUST00000121435.2 |
Gm12216
|
predicted gene 12216 |
| chr14_+_53061814 | 0.06 |
ENSMUST00000103648.4
|
Trav11d
|
T cell receptor alpha variable 11D |
| chrX_+_111513971 | 0.06 |
ENSMUST00000071814.13
|
Zfp711
|
zinc finger protein 711 |
| chr1_+_175526152 | 0.06 |
ENSMUST00000094288.10
ENSMUST00000171939.8 |
Wdr64
|
WD repeat domain 64 |
| chr14_-_55909314 | 0.06 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr12_+_17740831 | 0.05 |
ENSMUST00000071858.5
|
Hpcal1
|
hippocalcin-like 1 |
| chr5_-_120887864 | 0.05 |
ENSMUST00000053909.13
ENSMUST00000081491.13 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr13_+_83723255 | 0.05 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chrX_+_36528991 | 0.05 |
ENSMUST00000115194.5
|
Rhox4a
|
reproductive homeobox 4A |
| chr6_-_91784299 | 0.05 |
ENSMUST00000159684.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chrX_+_13527592 | 0.05 |
ENSMUST00000053659.2
|
Gpr82
|
G protein-coupled receptor 82 |
| chr18_+_44096300 | 0.05 |
ENSMUST00000069245.8
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr6_-_28133324 | 0.05 |
ENSMUST00000131897.2
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr17_-_41225241 | 0.05 |
ENSMUST00000166343.3
|
Glyatl3
|
glycine-N-acyltransferase-like 3 |
| chr2_+_111084861 | 0.05 |
ENSMUST00000218065.2
|
Olfr1276
|
olfactory receptor 1276 |
| chr14_-_55909527 | 0.05 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr18_+_34675366 | 0.05 |
ENSMUST00000012426.3
|
Wnt8a
|
wingless-type MMTV integration site family, member 8A |
| chr6_-_97037366 | 0.05 |
ENSMUST00000089295.6
|
Tafa4
|
TAFA chemokine like family member 4 |
| chrX_+_36739065 | 0.05 |
ENSMUST00000089075.6
|
Rhox4e
|
reproductive homeobox 4E |
| chr2_+_154390808 | 0.05 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chrX_-_72158969 | 0.05 |
ENSMUST00000114520.2
|
Xlr5a
|
X-linked lymphocyte-regulated 5A |
| chr2_+_89842475 | 0.05 |
ENSMUST00000214382.2
ENSMUST00000217065.3 |
Olfr1263
|
olfactory receptor 1263 |
| chr2_-_36752671 | 0.05 |
ENSMUST00000213676.2
ENSMUST00000215137.2 |
Olfr351
|
olfactory receptor 351 |
| chrX_+_72194740 | 0.05 |
ENSMUST00000097221.4
|
Xlr5b
|
X-linked lymphocyte-regulated 5B |
| chr14_-_79461316 | 0.05 |
ENSMUST00000040802.5
|
Zfp957
|
zinc finger protein 957 |
| chr7_+_54485336 | 0.04 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr17_+_70829144 | 0.04 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
| chr5_+_94796241 | 0.04 |
ENSMUST00000190001.7
|
Gm6205
|
predicted gene 6205 |
| chr18_+_36431732 | 0.04 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr19_+_5497575 | 0.04 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr5_+_94487608 | 0.04 |
ENSMUST00000097477.7
|
AA792892
|
expressed sequence AA792892 |
| chr3_+_96604415 | 0.04 |
ENSMUST00000107077.4
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr6_-_28133128 | 0.04 |
ENSMUST00000132755.2
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr7_-_34512101 | 0.04 |
ENSMUST00000078686.8
ENSMUST00000205259.2 ENSMUST00000154629.3 |
Chst8
|
carbohydrate sulfotransferase 8 |
| chr2_+_131792931 | 0.04 |
ENSMUST00000110169.2
|
Prnd
|
prion like protein doppel |
| chr6_-_91784405 | 0.04 |
ENSMUST00000162300.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chrX_-_72334121 | 0.04 |
ENSMUST00000088450.4
|
Xlr5c
|
X-linked lymphocyte-regulated 5C |
| chr6_+_30568366 | 0.04 |
ENSMUST00000049251.6
|
Cpa4
|
carboxypeptidase A4 |
| chr2_-_112089627 | 0.04 |
ENSMUST00000043970.2
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr14_+_53081922 | 0.04 |
ENSMUST00000181360.3
ENSMUST00000183652.2 |
Trav12d-1
|
T cell receptor alpha variable 12D-1 |
| chr2_+_140012560 | 0.04 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr12_+_87649458 | 0.04 |
ENSMUST00000222112.2
ENSMUST00000095509.6 ENSMUST00000222332.2 |
Oog1
|
oogenesin 1 |
| chr2_-_85027041 | 0.04 |
ENSMUST00000099930.9
ENSMUST00000111601.2 |
Lrrc55
|
leucine rich repeat containing 55 |
| chr6_+_96090127 | 0.04 |
ENSMUST00000122120.8
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr11_+_68792364 | 0.04 |
ENSMUST00000073471.13
ENSMUST00000101014.9 ENSMUST00000128952.8 ENSMUST00000167436.3 |
Rpl26
|
ribosomal protein L26 |
| chrX_-_142716200 | 0.04 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr6_-_69394425 | 0.04 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr5_+_24890810 | 0.03 |
ENSMUST00000068825.8
|
Nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr8_+_46080840 | 0.03 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr3_+_88536805 | 0.03 |
ENSMUST00000175745.2
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr12_+_87920850 | 0.03 |
ENSMUST00000180803.8
ENSMUST00000180707.8 ENSMUST00000181347.2 |
Gm2042
|
predicted gene 2042 |
| chr8_-_33374825 | 0.03 |
ENSMUST00000238791.2
|
Nrg1
|
neuregulin 1 |
| chr3_-_64317478 | 0.03 |
ENSMUST00000170280.3
|
Vmn2r4
|
vomeronasal 2, receptor 4 |
| chr1_+_55170411 | 0.03 |
ENSMUST00000027122.14
|
Mob4
|
MOB family member 4, phocein |
| chr17_+_70829050 | 0.03 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr7_+_103064279 | 0.03 |
ENSMUST00000106892.2
|
Usp17lc
|
ubiquitin specific peptidase 17-like C |
| chr3_-_108108113 | 0.02 |
ENSMUST00000106655.2
ENSMUST00000065664.7 |
Cyb561d1
|
cytochrome b-561 domain containing 1 |
| chr11_+_49213569 | 0.02 |
ENSMUST00000215671.2
|
Olfr1391
|
olfactory receptor 1391 |
| chr12_+_87921198 | 0.02 |
ENSMUST00000110145.12
ENSMUST00000181843.2 ENSMUST00000180706.8 ENSMUST00000181394.8 ENSMUST00000181326.8 ENSMUST00000181300.2 |
Gm2042
|
predicted gene 2042 |
| chr8_-_3904309 | 0.02 |
ENSMUST00000033888.5
|
Cd209e
|
CD209e antigen |
| chr6_+_142359099 | 0.02 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr14_+_53878403 | 0.02 |
ENSMUST00000184874.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr14_+_53853772 | 0.02 |
ENSMUST00000180972.3
|
Trav12-2
|
T cell receptor alpha variable 12-2 |
| chr10_-_33662700 | 0.02 |
ENSMUST00000223295.2
|
Sult3a2
|
sulfotransferase family 3A, member 2 |
| chr2_+_174485327 | 0.02 |
ENSMUST00000059452.6
|
Zfp831
|
zinc finger protein 831 |
| chr7_-_108251113 | 0.02 |
ENSMUST00000213756.2
ENSMUST00000215075.2 ENSMUST00000214861.2 |
Olfr509
|
olfactory receptor 509 |
| chr15_-_98796373 | 0.02 |
ENSMUST00000229775.2
ENSMUST00000023737.6 |
Dhh
|
desert hedgehog |
| chr4_+_62443606 | 0.02 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr6_+_41107047 | 0.02 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr2_+_88217406 | 0.02 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178 |
| chr6_+_29398919 | 0.01 |
ENSMUST00000181464.8
ENSMUST00000180829.8 |
Ccdc136
|
coiled-coil domain containing 136 |
| chr17_+_70828649 | 0.01 |
ENSMUST00000233283.2
|
Dlgap1
|
DLG associated protein 1 |
| chr6_-_122587005 | 0.01 |
ENSMUST00000032211.5
|
Gdf3
|
growth differentiation factor 3 |
| chr19_-_8196196 | 0.01 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr13_+_83672965 | 0.01 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr10_-_4338032 | 0.01 |
ENSMUST00000100078.10
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr12_+_38831093 | 0.01 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr11_-_101490011 | 0.01 |
ENSMUST00000209862.2
ENSMUST00000154147.2 |
Gm11634
|
predicted gene 11634 |
| chr1_+_85577766 | 0.01 |
ENSMUST00000066427.11
|
Sp100
|
nuclear antigen Sp100 |
| chr12_+_38830812 | 0.01 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr3_+_153549846 | 0.01 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr12_-_115083839 | 0.01 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr9_-_39618413 | 0.00 |
ENSMUST00000215192.2
|
Olfr149
|
olfactory receptor 149 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.1 | 0.4 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.1 | 2.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.2 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.1 | 0.2 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.2 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.0 | 0.0 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.0 | 0.1 | GO:0090370 | STAT protein import into nucleus(GO:0007262) negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |