Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
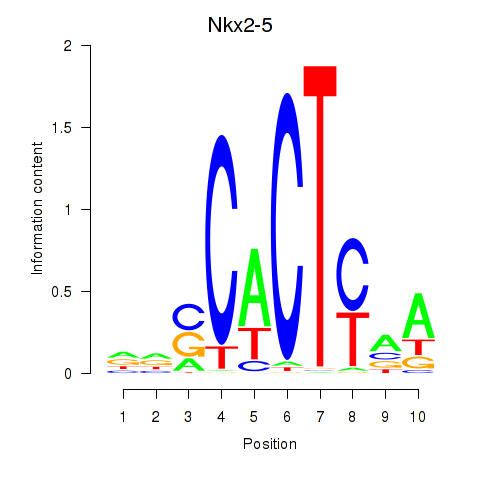
Results for Nkx2-5
Z-value: 1.16
Transcription factors associated with Nkx2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-5
|
ENSMUSG00000015579.6 | Nkx2-5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-5 | mm39_v1_chr17_-_27060539_27060565 | 0.23 | 1.8e-01 | Click! |
Activity profile of Nkx2-5 motif
Sorted Z-values of Nkx2-5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_30639217 | 21.20 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr6_+_41279199 | 18.05 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr6_+_30541581 | 11.44 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr4_-_137137088 | 10.23 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr6_-_136899167 | 5.90 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr1_+_74752710 | 4.77 |
ENSMUST00000027356.7
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr6_-_5193816 | 4.06 |
ENSMUST00000002663.12
|
Pon1
|
paraoxonase 1 |
| chr11_-_5900019 | 3.00 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr3_-_107145968 | 2.05 |
ENSMUST00000197758.5
|
Prok1
|
prokineticin 1 |
| chr8_+_107877252 | 1.98 |
ENSMUST00000034400.5
|
Cyb5b
|
cytochrome b5 type B |
| chr5_+_90708962 | 1.90 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr9_+_54824547 | 1.88 |
ENSMUST00000039742.8
|
Hykk
|
hydroxylysine kinase 1 |
| chr7_-_48493388 | 1.87 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr15_-_54783357 | 1.62 |
ENSMUST00000167541.3
ENSMUST00000171545.9 ENSMUST00000041591.16 ENSMUST00000173516.8 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr14_-_45767575 | 1.57 |
ENSMUST00000045905.15
|
Fermt2
|
fermitin family member 2 |
| chr9_+_106324952 | 1.46 |
ENSMUST00000215475.2
ENSMUST00000187106.7 ENSMUST00000190167.7 |
Abhd14b
|
abhydrolase domain containing 14b |
| chr2_+_22959223 | 1.17 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr11_-_96807192 | 1.15 |
ENSMUST00000144731.8
ENSMUST00000127048.8 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr7_-_126496534 | 1.05 |
ENSMUST00000120007.8
|
Tmem219
|
transmembrane protein 219 |
| chr12_+_8062331 | 0.98 |
ENSMUST00000171239.2
|
Apob
|
apolipoprotein B |
| chr11_+_68858942 | 0.97 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chrX_-_154123743 | 0.96 |
ENSMUST00000130349.3
|
Prdx4
|
peroxiredoxin 4 |
| chr9_-_50657800 | 0.95 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr1_-_66856695 | 0.94 |
ENSMUST00000068168.10
ENSMUST00000113987.2 |
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr5_+_28370687 | 0.85 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr2_+_127112127 | 0.81 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr9_-_53447908 | 0.81 |
ENSMUST00000150244.2
|
Atm
|
ataxia telangiectasia mutated |
| chr9_+_53212871 | 0.79 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr9_+_44684248 | 0.78 |
ENSMUST00000128150.8
|
Ift46
|
intraflagellar transport 46 |
| chr5_-_31448370 | 0.78 |
ENSMUST00000041565.11
ENSMUST00000201809.2 |
Ift172
|
intraflagellar transport 172 |
| chr9_+_44684285 | 0.72 |
ENSMUST00000239014.2
|
Ift46
|
intraflagellar transport 46 |
| chr3_-_57483330 | 0.70 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr17_-_56916771 | 0.69 |
ENSMUST00000052832.6
|
Micos13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chrX_+_41149264 | 0.68 |
ENSMUST00000224454.2
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr9_+_44684450 | 0.60 |
ENSMUST00000238800.2
ENSMUST00000147559.8 |
Ift46
|
intraflagellar transport 46 |
| chr16_+_43993599 | 0.59 |
ENSMUST00000119746.8
ENSMUST00000088356.10 ENSMUST00000169582.3 |
Usf3
|
upstream transcription factor family member 3 |
| chr9_+_44684324 | 0.58 |
ENSMUST00000214854.3
ENSMUST00000125877.8 |
Ift46
|
intraflagellar transport 46 |
| chr7_+_28869770 | 0.56 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr9_+_44684206 | 0.54 |
ENSMUST00000002099.11
|
Ift46
|
intraflagellar transport 46 |
| chr6_+_115830431 | 0.52 |
ENSMUST00000112925.8
ENSMUST00000038234.13 ENSMUST00000112923.7 |
Ift122
|
intraflagellar transport 122 |
| chr3_-_49711765 | 0.51 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr14_+_21550921 | 0.51 |
ENSMUST00000182964.3
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr14_+_53743184 | 0.50 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr2_-_173117936 | 0.49 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr2_+_32498716 | 0.48 |
ENSMUST00000129165.2
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr2_-_121287174 | 0.47 |
ENSMUST00000110613.9
ENSMUST00000056312.10 |
Serinc4
|
serine incorporator 4 |
| chrX_-_110372800 | 0.47 |
ENSMUST00000137712.9
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr6_-_29380423 | 0.43 |
ENSMUST00000147483.3
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr17_+_48037758 | 0.41 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr11_+_115865535 | 0.41 |
ENSMUST00000021107.14
ENSMUST00000169928.8 ENSMUST00000106461.8 |
Itgb4
|
integrin beta 4 |
| chr7_+_28869629 | 0.41 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr8_-_95839963 | 0.38 |
ENSMUST00000213004.2
|
Kifc3
|
kinesin family member C3 |
| chr11_-_78441584 | 0.37 |
ENSMUST00000103242.5
|
Tmem97
|
transmembrane protein 97 |
| chr9_-_119852624 | 0.36 |
ENSMUST00000111635.4
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr9_+_121245036 | 0.35 |
ENSMUST00000211187.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr11_+_64326141 | 0.35 |
ENSMUST00000058652.6
|
Hs3st3a1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr14_-_63509131 | 0.35 |
ENSMUST00000132122.2
|
Gata4
|
GATA binding protein 4 |
| chr5_-_71815318 | 0.34 |
ENSMUST00000199357.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr6_-_29380467 | 0.33 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr3_-_57483175 | 0.32 |
ENSMUST00000029380.14
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr4_+_137321451 | 0.31 |
ENSMUST00000105840.8
ENSMUST00000105839.8 ENSMUST00000055131.13 ENSMUST00000105838.8 |
Usp48
|
ubiquitin specific peptidase 48 |
| chr2_-_73722874 | 0.31 |
ENSMUST00000136958.8
ENSMUST00000112010.9 ENSMUST00000128531.8 ENSMUST00000112017.8 |
Atf2
|
activating transcription factor 2 |
| chr1_-_170755109 | 0.30 |
ENSMUST00000162136.2
ENSMUST00000162887.2 |
Fcrla
|
Fc receptor-like A |
| chr2_-_73722932 | 0.29 |
ENSMUST00000154456.8
ENSMUST00000090802.11 ENSMUST00000055833.12 ENSMUST00000112007.8 ENSMUST00000112016.9 |
Atf2
|
activating transcription factor 2 |
| chr12_+_84363876 | 0.29 |
ENSMUST00000222258.2
ENSMUST00000222832.2 ENSMUST00000220931.2 |
Zfp410
|
zinc finger protein 410 |
| chr2_+_173918089 | 0.29 |
ENSMUST00000155000.8
ENSMUST00000134876.8 ENSMUST00000147038.8 |
Stx16
|
syntaxin 16 |
| chrM_+_8603 | 0.29 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr5_+_71815382 | 0.27 |
ENSMUST00000199967.5
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr6_+_14901343 | 0.27 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr1_+_174257996 | 0.27 |
ENSMUST00000053178.5
|
Olfr414
|
olfactory receptor 414 |
| chr5_-_8417982 | 0.26 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr10_-_60667161 | 0.26 |
ENSMUST00000218637.2
|
Unc5b
|
unc-5 netrin receptor B |
| chrX_+_113384297 | 0.24 |
ENSMUST00000133447.2
|
Klhl4
|
kelch-like 4 |
| chr6_+_14901439 | 0.23 |
ENSMUST00000128567.8
|
Foxp2
|
forkhead box P2 |
| chr17_+_27152276 | 0.22 |
ENSMUST00000237412.2
|
Phf1
|
PHD finger protein 1 |
| chr8_-_84301386 | 0.21 |
ENSMUST00000238861.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr15_-_79212400 | 0.21 |
ENSMUST00000173163.8
ENSMUST00000047816.15 ENSMUST00000172403.9 ENSMUST00000173632.8 |
Pla2g6
|
phospholipase A2, group VI |
| chr8_-_84301457 | 0.20 |
ENSMUST00000238997.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr11_-_97909134 | 0.20 |
ENSMUST00000107561.9
|
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr12_-_115611981 | 0.17 |
ENSMUST00000103540.3
ENSMUST00000199266.2 |
Ighv8-12
|
immunoglobulin heavy variable V8-12 |
| chr11_-_99907030 | 0.17 |
ENSMUST00000018399.3
|
Krt33a
|
keratin 33A |
| chrX_-_110372733 | 0.17 |
ENSMUST00000065976.12
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr6_-_41535322 | 0.17 |
ENSMUST00000193003.2
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr6_-_41535292 | 0.15 |
ENSMUST00000103300.3
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr1_-_133589020 | 0.15 |
ENSMUST00000193504.6
ENSMUST00000195067.2 ENSMUST00000191896.6 ENSMUST00000194668.6 ENSMUST00000195424.6 ENSMUST00000179598.4 ENSMUST00000027736.13 |
Zc3h11a
Gm38394
Zc3h11a
|
zinc finger CCCH type containing 11A predicted gene, 38394 zinc finger CCCH type containing 11A |
| chrX_-_140508177 | 0.14 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr9_-_10904714 | 0.14 |
ENSMUST00000162484.8
ENSMUST00000160216.8 |
Cntn5
|
contactin 5 |
| chr11_+_67061908 | 0.14 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr4_+_85123654 | 0.13 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr8_+_114369838 | 0.13 |
ENSMUST00000095173.3
ENSMUST00000034219.12 ENSMUST00000212269.2 |
Syce1l
|
synaptonemal complex central element protein 1 like |
| chr5_+_124466146 | 0.13 |
ENSMUST00000111477.2
ENSMUST00000077376.3 |
2810006K23Rik
|
RIKEN cDNA 2810006K23 gene |
| chr11_+_87457479 | 0.13 |
ENSMUST00000239011.2
|
Septin4
|
septin 4 |
| chr8_-_107775204 | 0.12 |
ENSMUST00000055316.10
|
Pdf
|
peptide deformylase (mitochondrial) |
| chr15_-_79212323 | 0.12 |
ENSMUST00000166977.9
|
Pla2g6
|
phospholipase A2, group VI |
| chr3_-_63806794 | 0.12 |
ENSMUST00000161052.3
ENSMUST00000159188.2 ENSMUST00000177143.8 |
Plch1
|
phospholipase C, eta 1 |
| chr14_-_4506874 | 0.12 |
ENSMUST00000224934.2
|
Thrb
|
thyroid hormone receptor beta |
| chr7_-_79381911 | 0.10 |
ENSMUST00000178257.3
|
Plin1
|
perilipin 1 |
| chr12_-_114186874 | 0.10 |
ENSMUST00000103477.4
ENSMUST00000192499.3 |
Ighv7-4
|
immunoglobulin heavy variable 7-4 |
| chr11_+_67061837 | 0.10 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr15_-_5273645 | 0.10 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr3_-_129597679 | 0.09 |
ENSMUST00000185462.7
ENSMUST00000179187.2 |
Lrit3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr14_+_20724378 | 0.09 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr15_-_5273659 | 0.09 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr10_+_60235628 | 0.09 |
ENSMUST00000218974.2
|
Gm17455
|
predicted gene, 17455 |
| chr10_-_114638202 | 0.07 |
ENSMUST00000239411.2
|
Trhde
|
TRH-degrading enzyme |
| chr12_+_103281188 | 0.07 |
ENSMUST00000189885.2
ENSMUST00000179363.2 |
Fam181a
|
family with sequence similarity 181, member A |
| chr7_-_4909515 | 0.07 |
ENSMUST00000210663.2
|
Gm36210
|
predicted gene, 36210 |
| chr12_+_37930661 | 0.07 |
ENSMUST00000040500.9
|
Dgkb
|
diacylglycerol kinase, beta |
| chr7_+_114318746 | 0.06 |
ENSMUST00000182044.2
|
Calcb
|
calcitonin-related polypeptide, beta |
| chr6_-_52222776 | 0.06 |
ENSMUST00000048026.10
|
Hoxa11
|
homeobox A11 |
| chr6_-_90013823 | 0.04 |
ENSMUST00000073415.2
|
Vmn1r48
|
vomeronasal 1 receptor 48 |
| chr11_-_101877832 | 0.04 |
ENSMUST00000107173.9
ENSMUST00000107172.8 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr12_-_115531211 | 0.04 |
ENSMUST00000103536.4
|
Ighv8-11
|
immunoglobulin heavy variable V8-11 |
| chr15_-_99355593 | 0.03 |
ENSMUST00000161948.2
|
Nckap5l
|
NCK-associated protein 5-like |
| chr6_-_28421678 | 0.03 |
ENSMUST00000090511.4
|
Gcc1
|
golgi coiled coil 1 |
| chr11_+_87457544 | 0.03 |
ENSMUST00000060360.7
|
Septin4
|
septin 4 |
| chr11_+_73125118 | 0.03 |
ENSMUST00000102526.9
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chrX_-_164110372 | 0.03 |
ENSMUST00000058787.9
|
Glra2
|
glycine receptor, alpha 2 subunit |
| chr12_-_114286421 | 0.02 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr10_+_60236532 | 0.02 |
ENSMUST00000218360.2
|
Gm17455
|
predicted gene, 17455 |
| chr10_-_116732813 | 0.02 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr14_+_47238772 | 0.02 |
ENSMUST00000125113.9
|
Samd4
|
sterile alpha motif domain containing 4 |
| chr8_+_63404395 | 0.01 |
ENSMUST00000119068.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr14_-_78326435 | 0.01 |
ENSMUST00000118785.3
ENSMUST00000066437.5 |
Fam216b
|
family with sequence similarity 216, member B |
| chr3_-_89067462 | 0.01 |
ENSMUST00000029686.4
|
Hcn3
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
| chr14_+_52131067 | 0.01 |
ENSMUST00000047726.12
|
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr16_+_33504908 | 0.00 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.6 | 1.9 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.6 | 3.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.3 | 0.8 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.2 | 1.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 1.0 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.2 | 0.5 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 0.7 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.6 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 1.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.4 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.0 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.3 | GO:0060464 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 0.1 | 0.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 3.2 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 1.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 18.1 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.0 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 15.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 0.2 | GO:1904348 | negative regulation of gastro-intestinal system smooth muscle contraction(GO:1904305) negative regulation of small intestine smooth muscle contraction(GO:1904348) |
| 0.1 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.3 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 1.6 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.8 | GO:0061525 | hindgut development(GO:0061525) left/right axis specification(GO:0070986) |
| 0.0 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.5 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.4 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 2.0 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 29.9 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0051324 | M phase(GO:0000279) meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.0 | 0.8 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.3 | 4.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 0.8 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 2.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 1.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 54.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 7.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 32.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.0 | 4.1 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.3 | 1.6 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 3.0 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.3 | 0.8 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.3 | 1.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 28.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.0 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 4.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.0 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 1.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.0 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 2.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 10.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |