Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
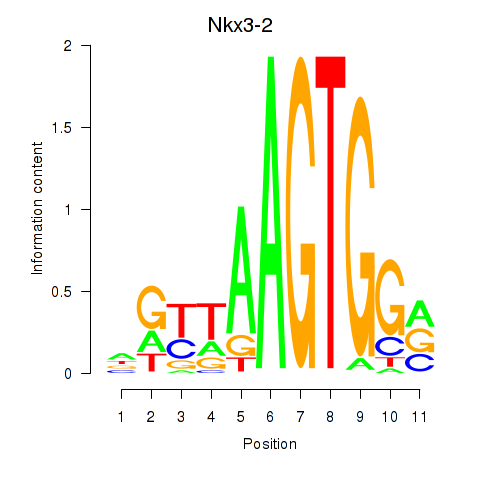
Results for Nkx3-2
Z-value: 2.43
Transcription factors associated with Nkx3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx3-2
|
ENSMUSG00000049691.9 | Nkx3-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx3-2 | mm39_v1_chr5_-_41921834_41921844 | 0.30 | 7.3e-02 | Click! |
Activity profile of Nkx3-2 motif
Sorted Z-values of Nkx3-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx3-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90638580 | 19.88 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr8_+_81220410 | 14.40 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr14_-_20319242 | 10.07 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr8_-_73059104 | 8.95 |
ENSMUST00000075602.8
|
Gm10282
|
predicted pseudogene 10282 |
| chr11_+_70529944 | 8.80 |
ENSMUST00000055184.7
ENSMUST00000108551.3 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr2_-_165242307 | 8.47 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr7_+_142559414 | 7.89 |
ENSMUST00000082008.12
ENSMUST00000105925.8 ENSMUST00000105924.8 |
Tspan32
|
tetraspanin 32 |
| chr1_-_75110511 | 7.49 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr14_-_79539063 | 7.45 |
ENSMUST00000022595.8
|
Rgcc
|
regulator of cell cycle |
| chr7_+_142559375 | 6.97 |
ENSMUST00000075172.12
ENSMUST00000105923.8 |
Tspan32
|
tetraspanin 32 |
| chrX_-_135104589 | 6.56 |
ENSMUST00000066819.11
|
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr7_+_78563964 | 6.23 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr10_+_79824418 | 5.79 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr1_+_91294133 | 5.66 |
ENSMUST00000086861.12
|
Erfe
|
erythroferrone |
| chr15_-_103161237 | 5.58 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr7_-_110581652 | 5.56 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr7_-_103502404 | 5.29 |
ENSMUST00000033229.5
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr15_+_34306812 | 5.29 |
ENSMUST00000226766.2
ENSMUST00000163455.9 ENSMUST00000022947.7 ENSMUST00000228570.2 ENSMUST00000227759.2 |
Matn2
|
matrilin 2 |
| chr11_+_120653613 | 4.88 |
ENSMUST00000105046.4
|
Hmga1b
|
high mobility group AT-hook 1B |
| chr9_+_110856425 | 4.78 |
ENSMUST00000199313.2
|
Ltf
|
lactotransferrin |
| chr10_+_127350820 | 4.62 |
ENSMUST00000035735.11
|
Ndufa4l2
|
Ndufa4, mitochondrial complex associated like 2 |
| chr4_-_140501507 | 4.46 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr8_-_86107593 | 4.44 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr15_-_100567377 | 4.35 |
ENSMUST00000182814.8
ENSMUST00000238935.2 ENSMUST00000182068.8 ENSMUST00000182574.2 ENSMUST00000182775.8 |
Bin2
|
bridging integrator 2 |
| chr19_-_24533183 | 4.23 |
ENSMUST00000112673.9
ENSMUST00000025800.15 |
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta |
| chr4_+_130253925 | 4.21 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr7_+_142559475 | 3.89 |
ENSMUST00000143512.3
|
Tspan32
|
tetraspanin 32 |
| chrX_-_135104386 | 3.73 |
ENSMUST00000151592.8
ENSMUST00000131510.2 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr10_+_79852487 | 3.60 |
ENSMUST00000099501.10
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr8_-_4829519 | 3.50 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr3_-_67371161 | 3.43 |
ENSMUST00000058981.3
|
Lxn
|
latexin |
| chr18_+_50186349 | 3.42 |
ENSMUST00000148159.3
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr6_+_116327853 | 3.34 |
ENSMUST00000140884.8
ENSMUST00000129170.8 |
Marchf8
|
membrane associated ring-CH-type finger 8 |
| chr8_+_46338498 | 3.22 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr4_+_63477018 | 3.16 |
ENSMUST00000077709.11
|
Tmem268
|
transmembrane protein 268 |
| chr7_+_127503251 | 3.16 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr2_-_34645241 | 3.15 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr11_+_117673107 | 3.07 |
ENSMUST00000050874.14
ENSMUST00000106334.9 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr9_+_48073296 | 3.05 |
ENSMUST00000216998.2
ENSMUST00000215780.2 |
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr8_-_123278054 | 3.04 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr1_+_136395673 | 3.01 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr9_-_62719208 | 2.92 |
ENSMUST00000034775.10
|
Fem1b
|
fem 1 homolog b |
| chr1_+_160898283 | 2.91 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr7_-_29979758 | 2.83 |
ENSMUST00000108190.8
|
Wdr62
|
WD repeat domain 62 |
| chr1_-_181670599 | 2.83 |
ENSMUST00000193030.6
|
Lbr
|
lamin B receptor |
| chr15_-_102112657 | 2.78 |
ENSMUST00000231030.2
ENSMUST00000230687.2 ENSMUST00000229514.2 ENSMUST00000229345.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr5_+_64250268 | 2.68 |
ENSMUST00000087324.7
|
Pgm2
|
phosphoglucomutase 2 |
| chr9_-_45180433 | 2.67 |
ENSMUST00000176222.2
ENSMUST00000034594.16 |
Il10ra
|
interleukin 10 receptor, alpha |
| chr2_+_32253016 | 2.66 |
ENSMUST00000132028.8
ENSMUST00000136079.8 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr8_+_27575611 | 2.65 |
ENSMUST00000178514.8
ENSMUST00000033876.14 |
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr1_-_171061902 | 2.64 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr15_-_100567412 | 2.63 |
ENSMUST00000183211.8
|
Bin2
|
bridging integrator 2 |
| chr7_+_97102411 | 2.63 |
ENSMUST00000121987.3
ENSMUST00000050732.14 ENSMUST00000205577.2 ENSMUST00000206279.2 |
Kctd14
|
potassium channel tetramerisation domain containing 14 |
| chr19_-_7218512 | 2.53 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr19_-_7218363 | 2.49 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr5_+_30360246 | 2.48 |
ENSMUST00000026841.15
ENSMUST00000123980.8 ENSMUST00000114783.6 ENSMUST00000114786.8 |
Hadhb
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr18_+_23885390 | 2.45 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr17_-_71833752 | 2.40 |
ENSMUST00000232863.2
ENSMUST00000024851.10 |
Ndc80
|
NDC80 kinetochore complex component |
| chr10_-_128755127 | 2.38 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr2_-_156833932 | 2.35 |
ENSMUST00000109558.2
ENSMUST00000069600.13 ENSMUST00000072298.13 |
Ndrg3
|
N-myc downstream regulated gene 3 |
| chr2_+_32253692 | 2.35 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr3_+_95071617 | 2.33 |
ENSMUST00000168321.8
ENSMUST00000107217.6 ENSMUST00000202315.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr16_+_22710785 | 2.23 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr8_+_46338557 | 2.19 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr2_-_35351259 | 2.14 |
ENSMUST00000113001.9
ENSMUST00000113002.9 |
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chrX_+_164953444 | 2.10 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr18_-_35781422 | 2.09 |
ENSMUST00000237462.2
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chrX_-_7814087 | 2.06 |
ENSMUST00000115642.8
ENSMUST00000033501.15 ENSMUST00000145675.8 |
Hdac6
|
histone deacetylase 6 |
| chr11_+_117673198 | 2.01 |
ENSMUST00000117781.8
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr2_+_32253204 | 1.99 |
ENSMUST00000048964.14
ENSMUST00000113332.8 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr7_-_125090540 | 1.99 |
ENSMUST00000138616.3
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr17_+_34812361 | 1.96 |
ENSMUST00000174532.2
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr16_+_57173456 | 1.94 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr16_+_4704103 | 1.93 |
ENSMUST00000023159.10
ENSMUST00000070658.16 ENSMUST00000229038.2 |
Mgrn1
|
mahogunin, ring finger 1 |
| chrX_-_101295787 | 1.92 |
ENSMUST00000050551.10
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr4_-_56802266 | 1.92 |
ENSMUST00000030140.3
|
Elp1
|
elongator complex protein 1 |
| chrX_+_149829131 | 1.90 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chrX_+_139857640 | 1.90 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr16_-_18884431 | 1.89 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr3_-_81981946 | 1.83 |
ENSMUST00000029635.14
ENSMUST00000193597.2 |
Gucy1b1
|
guanylate cyclase 1, soluble, beta 1 |
| chr16_+_4704166 | 1.83 |
ENSMUST00000230990.2
|
Mgrn1
|
mahogunin, ring finger 1 |
| chr15_-_100585789 | 1.81 |
ENSMUST00000023775.9
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr6_-_72357398 | 1.78 |
ENSMUST00000101285.10
ENSMUST00000074231.6 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr2_+_24276616 | 1.77 |
ENSMUST00000166388.2
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr9_-_107971640 | 1.77 |
ENSMUST00000081309.13
ENSMUST00000191985.2 |
Apeh
|
acylpeptide hydrolase |
| chr13_+_56757389 | 1.74 |
ENSMUST00000045173.10
|
Tgfbi
|
transforming growth factor, beta induced |
| chr19_+_45035942 | 1.74 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chr2_-_35351408 | 1.71 |
ENSMUST00000102794.8
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr7_-_125090757 | 1.70 |
ENSMUST00000033006.14
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr7_-_5016237 | 1.67 |
ENSMUST00000208944.2
|
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr5_-_110987441 | 1.66 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr13_-_24945423 | 1.66 |
ENSMUST00000176890.8
ENSMUST00000175689.8 |
Gmnn
|
geminin |
| chr9_-_56151334 | 1.66 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr16_+_22713593 | 1.61 |
ENSMUST00000232674.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr8_+_105066924 | 1.60 |
ENSMUST00000212081.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr19_+_45036037 | 1.57 |
ENSMUST00000062213.13
|
Sfxn3
|
sideroflexin 3 |
| chr4_+_149670889 | 1.56 |
ENSMUST00000105691.8
|
Clstn1
|
calsyntenin 1 |
| chr7_-_19093383 | 1.55 |
ENSMUST00000047036.10
|
Cd3eap
|
CD3E antigen, epsilon polypeptide associated protein |
| chrX_+_139857688 | 1.54 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr10_+_52109707 | 1.52 |
ENSMUST00000069004.14
ENSMUST00000218582.2 |
Dcbld1
|
discoidin, CUB and LCCL domain containing 1 |
| chr5_-_30360113 | 1.51 |
ENSMUST00000156859.3
|
Hadha
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha |
| chrX_-_100312629 | 1.51 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr1_+_135764092 | 1.49 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr7_+_127836502 | 1.45 |
ENSMUST00000044660.6
|
Armc5
|
armadillo repeat containing 5 |
| chr8_+_104828253 | 1.45 |
ENSMUST00000034339.10
|
Cdh5
|
cadherin 5 |
| chr2_+_29509704 | 1.42 |
ENSMUST00000095087.11
ENSMUST00000091146.12 ENSMUST00000102872.11 ENSMUST00000147755.10 |
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr5_-_113163339 | 1.42 |
ENSMUST00000197776.2
ENSMUST00000065167.9 |
Grk3
|
G protein-coupled receptor kinase 3 |
| chr1_-_192880260 | 1.42 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chrX_+_100317629 | 1.42 |
ENSMUST00000117706.8
|
Med12
|
mediator complex subunit 12 |
| chr11_-_117673008 | 1.40 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr2_+_121337226 | 1.39 |
ENSMUST00000099473.10
ENSMUST00000110602.9 |
Wdr76
|
WD repeat domain 76 |
| chr2_+_130266253 | 1.38 |
ENSMUST00000128994.8
ENSMUST00000028900.11 |
Vps16
|
VSP16 CORVET/HOPS core subunit |
| chr9_+_98445757 | 1.38 |
ENSMUST00000035033.7
|
Copb2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr13_-_100753419 | 1.36 |
ENSMUST00000168772.2
ENSMUST00000163163.9 ENSMUST00000022137.14 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr11_-_121119864 | 1.36 |
ENSMUST00000137299.8
|
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr5_-_124327776 | 1.34 |
ENSMUST00000159677.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr8_+_75940572 | 1.32 |
ENSMUST00000139848.8
|
Rasd2
|
RASD family, member 2 |
| chr11_-_100510992 | 1.32 |
ENSMUST00000014339.15
ENSMUST00000239490.2 ENSMUST00000239410.2 |
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr3_+_89344006 | 1.28 |
ENSMUST00000038942.10
ENSMUST00000130858.8 ENSMUST00000146630.8 ENSMUST00000145753.2 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr5_+_115644727 | 1.26 |
ENSMUST00000067268.15
ENSMUST00000086523.7 ENSMUST00000212819.3 |
Pxn
|
paxillin |
| chr6_+_83112793 | 1.26 |
ENSMUST00000065512.11
|
Rtkn
|
rhotekin |
| chr9_-_58220469 | 1.24 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr19_-_4927910 | 1.23 |
ENSMUST00000006626.5
|
Actn3
|
actinin alpha 3 |
| chr1_+_39940189 | 1.21 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr9_-_45895635 | 1.21 |
ENSMUST00000215427.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr11_-_78642480 | 1.20 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr4_+_56802338 | 1.20 |
ENSMUST00000045368.12
|
Abitram
|
actin binding transcription modulator |
| chr9_+_64024429 | 1.17 |
ENSMUST00000034969.14
|
Lctl
|
lactase-like |
| chr19_-_24178000 | 1.16 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr7_+_43000765 | 1.15 |
ENSMUST00000012798.14
ENSMUST00000122423.8 |
Siglecf
|
sialic acid binding Ig-like lectin F |
| chr11_+_43365103 | 1.15 |
ENSMUST00000173002.8
ENSMUST00000057679.10 |
C1qtnf2
|
C1q and tumor necrosis factor related protein 2 |
| chr8_-_112737952 | 1.15 |
ENSMUST00000034426.14
|
Kars
|
lysyl-tRNA synthetase |
| chr11_-_101877832 | 1.15 |
ENSMUST00000107173.9
ENSMUST00000107172.8 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr9_+_69902697 | 1.13 |
ENSMUST00000165389.8
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr19_+_45036220 | 1.12 |
ENSMUST00000084493.8
|
Sfxn3
|
sideroflexin 3 |
| chr11_+_72580823 | 1.12 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr16_+_20354225 | 1.11 |
ENSMUST00000090023.13
ENSMUST00000007216.9 ENSMUST00000232001.2 |
Ap2m1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr11_-_5787743 | 1.10 |
ENSMUST00000109837.8
|
Polm
|
polymerase (DNA directed), mu |
| chr7_+_140436870 | 1.09 |
ENSMUST00000026558.7
|
Ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr2_+_87696836 | 1.08 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr18_-_36648850 | 1.08 |
ENSMUST00000025363.7
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr8_+_4216556 | 1.08 |
ENSMUST00000239400.2
ENSMUST00000177053.8 ENSMUST00000176149.9 ENSMUST00000176072.9 ENSMUST00000176825.3 |
Evi5l
|
ecotropic viral integration site 5 like |
| chr7_+_122564859 | 1.07 |
ENSMUST00000148880.2
|
Rbbp6
|
retinoblastoma binding protein 6, ubiquitin ligase |
| chr8_-_112737898 | 1.04 |
ENSMUST00000093120.13
ENSMUST00000164470.3 ENSMUST00000211990.2 |
Kars
|
lysyl-tRNA synthetase |
| chr10_+_17931459 | 1.04 |
ENSMUST00000154718.8
ENSMUST00000126390.8 ENSMUST00000164556.8 ENSMUST00000150029.8 |
Reps1
|
RalBP1 associated Eps domain containing protein |
| chr8_-_123404811 | 1.03 |
ENSMUST00000006525.14
ENSMUST00000064674.13 |
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr2_+_158636727 | 1.02 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr4_-_40239700 | 1.01 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr9_-_44646487 | 1.01 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr11_+_82782938 | 1.00 |
ENSMUST00000018988.6
|
Fndc8
|
fibronectin type III domain containing 8 |
| chr8_-_94825556 | 0.99 |
ENSMUST00000034206.6
|
Bbs2
|
Bardet-Biedl syndrome 2 (human) |
| chr10_-_79936987 | 0.99 |
ENSMUST00000218630.2
|
Sbno2
|
strawberry notch 2 |
| chr3_-_95214102 | 0.98 |
ENSMUST00000107183.8
ENSMUST00000164406.8 ENSMUST00000123365.2 |
Anxa9
|
annexin A9 |
| chr4_-_58206596 | 0.96 |
ENSMUST00000042850.9
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chrX_-_7813792 | 0.95 |
ENSMUST00000133349.2
|
Hdac6
|
histone deacetylase 6 |
| chr2_+_128540822 | 0.94 |
ENSMUST00000014505.5
|
Mertk
|
MER proto-oncogene tyrosine kinase |
| chr7_+_43000838 | 0.93 |
ENSMUST00000206299.2
ENSMUST00000121494.2 |
Siglecf
|
sialic acid binding Ig-like lectin F |
| chr7_-_103778992 | 0.92 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr4_-_151129020 | 0.88 |
ENSMUST00000103204.11
|
Per3
|
period circadian clock 3 |
| chr7_-_44542098 | 0.87 |
ENSMUST00000003049.8
|
Med25
|
mediator complex subunit 25 |
| chr8_-_110292366 | 0.86 |
ENSMUST00000042601.9
|
Dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr15_-_102112159 | 0.86 |
ENSMUST00000229252.2
ENSMUST00000229770.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr13_-_113063890 | 0.85 |
ENSMUST00000022281.5
|
Mtrex
|
Mtr4 exosome RNA helicase |
| chr5_+_136112765 | 0.84 |
ENSMUST00000042135.14
|
Rasa4
|
RAS p21 protein activator 4 |
| chr11_-_94440025 | 0.84 |
ENSMUST00000040487.4
|
Rsad1
|
radical S-adenosyl methionine domain containing 1 |
| chr1_+_172168764 | 0.80 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr8_+_40876827 | 0.80 |
ENSMUST00000049389.11
ENSMUST00000128166.8 ENSMUST00000167766.2 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr13_+_43938251 | 0.78 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr7_-_109559671 | 0.78 |
ENSMUST00000080437.13
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr4_-_130253703 | 0.77 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr17_+_36134450 | 0.76 |
ENSMUST00000172846.2
|
Flot1
|
flotillin 1 |
| chr6_+_49344673 | 0.76 |
ENSMUST00000060561.15
ENSMUST00000121903.2 ENSMUST00000134786.2 |
Fam221a
|
family with sequence similarity 221, member A |
| chr8_-_57003828 | 0.72 |
ENSMUST00000134162.8
ENSMUST00000140107.8 ENSMUST00000040330.15 ENSMUST00000135337.8 |
Cep44
|
centrosomal protein 44 |
| chr17_-_43854530 | 0.71 |
ENSMUST00000178772.3
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr10_+_129153986 | 0.70 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr13_-_100753181 | 0.70 |
ENSMUST00000225754.2
|
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr9_-_107971729 | 0.69 |
ENSMUST00000193254.6
|
Apeh
|
acylpeptide hydrolase |
| chr11_-_100510423 | 0.69 |
ENSMUST00000137688.9
ENSMUST00000239389.2 ENSMUST00000155152.9 ENSMUST00000154972.9 ENSMUST00000239479.2 ENSMUST00000132886.3 |
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr8_-_69541852 | 0.68 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr7_-_109559593 | 0.68 |
ENSMUST00000106722.2
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr19_-_5438685 | 0.67 |
ENSMUST00000044207.5
|
Sart1
|
squamous cell carcinoma antigen recognized by T cells 1 |
| chr7_+_92390811 | 0.66 |
ENSMUST00000032879.15
|
Rab30
|
RAB30, member RAS oncogene family |
| chr18_+_74401322 | 0.66 |
ENSMUST00000224047.2
|
Mbd1
|
methyl-CpG binding domain protein 1 |
| chr3_-_95214443 | 0.66 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr11_+_49141410 | 0.65 |
ENSMUST00000129588.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr2_-_165129675 | 0.65 |
ENSMUST00000109299.8
ENSMUST00000109298.8 ENSMUST00000130393.2 ENSMUST00000017808.14 ENSMUST00000131409.2 ENSMUST00000156134.8 ENSMUST00000133961.8 |
Slc35c2
|
solute carrier family 35, member C2 |
| chr11_+_69886652 | 0.64 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr17_+_15616464 | 0.61 |
ENSMUST00000055352.8
|
Fam120b
|
family with sequence similarity 120, member B |
| chr6_+_83011154 | 0.61 |
ENSMUST00000000707.9
ENSMUST00000101257.4 |
Loxl3
|
lysyl oxidase-like 3 |
| chr11_-_32217547 | 0.58 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr11_+_117672902 | 0.58 |
ENSMUST00000127080.9
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr11_+_69886603 | 0.58 |
ENSMUST00000018716.10
|
Phf23
|
PHD finger protein 23 |
| chr16_+_21023505 | 0.57 |
ENSMUST00000006112.7
|
Ephb3
|
Eph receptor B3 |
| chr7_-_30855295 | 0.56 |
ENSMUST00000186723.3
|
Gramd1a
|
GRAM domain containing 1A |
| chrX_-_99670174 | 0.54 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chrX_-_99669507 | 0.54 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11 |
| chr11_-_109613040 | 0.53 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr4_+_156110621 | 0.53 |
ENSMUST00000103173.10
ENSMUST00000040274.13 ENSMUST00000122001.3 |
Tnfrsf18
|
tumor necrosis factor receptor superfamily, member 18 |
| chr10_-_13350106 | 0.51 |
ENSMUST00000105545.12
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr11_+_49141339 | 0.51 |
ENSMUST00000101293.11
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 2.1 | 8.5 | GO:0090290 | positive regulation of macrophage fusion(GO:0034241) positive regulation of osteoclast proliferation(GO:0090290) |
| 1.8 | 5.3 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 1.7 | 7.0 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 1.7 | 18.7 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 1.2 | 19.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.2 | 14.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 1.2 | 4.8 | GO:0042710 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.0 | 6.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.0 | 3.0 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 1.0 | 3.8 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.9 | 4.5 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.8 | 2.5 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.7 | 2.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.7 | 2.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.7 | 3.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.7 | 4.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.7 | 2.6 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.6 | 3.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 5.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.6 | 10.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.5 | 3.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.4 | 2.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.4 | 2.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.4 | 1.2 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.4 | 1.9 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.4 | 5.6 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.4 | 1.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.4 | 1.1 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.3 | 8.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 2.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 1.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.3 | 0.9 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.3 | 1.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.3 | 3.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.3 | 2.4 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.3 | 1.8 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.2 | 1.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 6.0 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 1.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.8 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) regulation of resting membrane potential(GO:0060075) |
| 0.2 | 7.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 5.7 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.2 | 0.2 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.2 | 5.8 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.2 | 1.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 0.9 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.2 | 1.1 | GO:0032439 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.2 | 1.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 1.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 5.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.3 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 0.6 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.2 | 1.4 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 1.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 3.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 0.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 1.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.4 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 3.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 3.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.8 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 1.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 2.4 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.6 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 2.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.7 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.1 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.8 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 | 1.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 1.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.0 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 1.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 1.1 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.1 | 0.5 | GO:0098735 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 2.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 4.2 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.1 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.1 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.1 | 2.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 1.4 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 | 3.4 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 1.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 1.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 4.2 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 1.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 1.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.7 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.8 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 1.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.9 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.0 | 3.5 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 1.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.6 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.6 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.0 | 1.2 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 3.8 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 4.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.1 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 1.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.7 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.9 | 2.6 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.8 | 4.8 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.7 | 5.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.6 | 2.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.5 | 3.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.4 | 3.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 8.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 5.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 4.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 2.1 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.2 | 1.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 7.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 5.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 2.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 1.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 2.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 6.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 7.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 6.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.8 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 1.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 3.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 11.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 4.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 12.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 12.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 2.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 3.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 3.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0061702 | inflammasome complex(GO:0061702) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 19.9 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 1.3 | 4.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.3 | 5.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 1.2 | 6.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.9 | 4.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.6 | 3.8 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.6 | 3.8 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.6 | 5.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.5 | 10.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.5 | 2.6 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.5 | 3.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.5 | 2.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.5 | 4.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.5 | 4.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 2.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 2.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.3 | 2.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 1.3 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.3 | 1.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 1.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 15.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 2.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 1.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 1.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 7.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 1.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 0.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 0.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 4.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 1.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 4.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 3.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 7.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 2.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 3.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 2.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.7 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.1 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 8.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 3.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 3.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 5.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 2.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 2.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 5.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 19.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 5.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 5.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 6.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.7 | 10.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 6.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 6.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 4.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 4.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 5.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |