Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr0b1
Z-value: 1.01
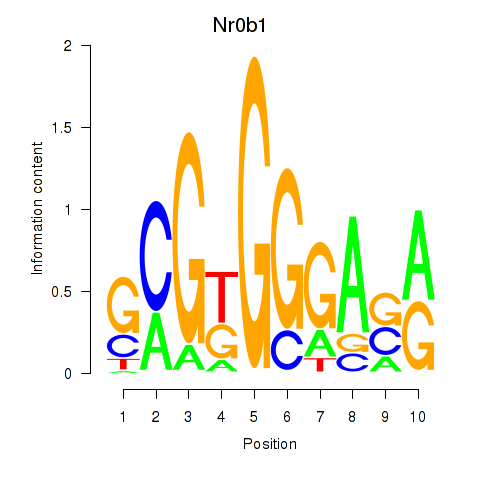
Transcription factors associated with Nr0b1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr0b1
|
ENSMUSG00000025056.5 | Nr0b1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | mm39_v1_chrX_+_85235370_85235388 | -0.38 | 2.4e-02 | Click! |
Activity profile of Nr0b1 motif
Sorted Z-values of Nr0b1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr0b1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_88015524 | 5.01 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr6_-_72212547 | 3.54 |
ENSMUST00000042646.8
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr12_-_104831335 | 3.36 |
ENSMUST00000109936.3
|
Clmn
|
calmin |
| chr16_-_20440005 | 2.70 |
ENSMUST00000052939.4
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr2_+_27567213 | 2.18 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr4_+_148215339 | 2.17 |
ENSMUST00000084129.9
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr11_-_69260203 | 2.15 |
ENSMUST00000092971.13
ENSMUST00000108661.8 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr15_-_85918378 | 2.12 |
ENSMUST00000016172.10
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr19_+_37423198 | 1.92 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr4_+_53826013 | 1.79 |
ENSMUST00000030127.13
|
Tmem38b
|
transmembrane protein 38B |
| chr10_-_128425519 | 1.61 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr8_-_71834543 | 1.60 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr2_+_157120946 | 1.57 |
ENSMUST00000116380.9
ENSMUST00000029171.6 |
Rpn2
|
ribophorin II |
| chr5_+_30972067 | 1.57 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr2_+_4722956 | 1.56 |
ENSMUST00000056914.7
|
Bend7
|
BEN domain containing 7 |
| chr10_-_127504416 | 1.36 |
ENSMUST00000129252.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr9_-_110571645 | 1.35 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr2_-_144173615 | 1.27 |
ENSMUST00000103171.10
|
Ovol2
|
ovo like zinc finger 2 |
| chr10_-_81186025 | 1.20 |
ENSMUST00000122993.8
|
Hmg20b
|
high mobility group 20B |
| chr10_-_81186137 | 1.18 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr10_-_81186222 | 1.11 |
ENSMUST00000020454.11
ENSMUST00000105324.9 ENSMUST00000154609.3 ENSMUST00000105323.8 |
Hmg20b
|
high mobility group 20B |
| chr8_-_71308040 | 1.10 |
ENSMUST00000212509.3
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr6_+_85164420 | 1.06 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chr8_-_71308229 | 1.05 |
ENSMUST00000212086.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr11_-_115503704 | 1.04 |
ENSMUST00000106506.8
|
Mif4gd
|
MIF4G domain containing |
| chr1_+_140173787 | 1.03 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr15_+_35371644 | 1.02 |
ENSMUST00000227455.2
|
Vps13b
|
vacuolar protein sorting 13B |
| chr19_+_23736205 | 1.01 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr4_+_116078874 | 0.99 |
ENSMUST00000106490.3
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr4_+_116078787 | 0.88 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr18_+_35963353 | 0.79 |
ENSMUST00000235169.2
|
Cxxc5
|
CXXC finger 5 |
| chr11_-_115503316 | 0.76 |
ENSMUST00000106507.9
|
Mif4gd
|
MIF4G domain containing |
| chr8_+_85807369 | 0.72 |
ENSMUST00000079764.14
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr11_-_69871320 | 0.71 |
ENSMUST00000143175.2
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr16_+_56942050 | 0.68 |
ENSMUST00000166897.3
|
Tomm70a
|
translocase of outer mitochondrial membrane 70A |
| chr8_+_106434901 | 0.67 |
ENSMUST00000013302.7
ENSMUST00000211852.2 |
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr19_-_23425757 | 0.66 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr7_+_45434755 | 0.65 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr17_+_88837540 | 0.65 |
ENSMUST00000038551.8
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr7_-_80020830 | 0.64 |
ENSMUST00000205436.2
ENSMUST00000098346.5 |
Man2a2
|
mannosidase 2, alpha 2 |
| chr17_-_23964807 | 0.63 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr5_-_108134869 | 0.63 |
ENSMUST00000145239.2
ENSMUST00000031198.11 |
Dipk1a
|
divergent protein kinase domain 1A |
| chr2_-_17735847 | 0.62 |
ENSMUST00000028080.12
|
Nebl
|
nebulette |
| chr2_+_81883566 | 0.58 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr14_-_61677258 | 0.56 |
ENSMUST00000022496.9
|
Kpna3
|
karyopherin (importin) alpha 3 |
| chr4_+_116078830 | 0.56 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr11_-_88608958 | 0.52 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_-_30560989 | 0.50 |
ENSMUST00000052700.6
|
Ffar1
|
free fatty acid receptor 1 |
| chr8_-_85807281 | 0.50 |
ENSMUST00000152785.8
|
Wdr83
|
WD repeat domain containing 83 |
| chr9_-_119812042 | 0.49 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr5_-_31350352 | 0.49 |
ENSMUST00000202758.4
ENSMUST00000114603.8 |
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr8_-_85807308 | 0.48 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr15_+_81695615 | 0.46 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chrX_-_37653396 | 0.46 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr8_+_56393488 | 0.43 |
ENSMUST00000000275.10
|
Glra3
|
glycine receptor, alpha 3 subunit |
| chr7_-_80020590 | 0.43 |
ENSMUST00000206212.2
|
Man2a2
|
mannosidase 2, alpha 2 |
| chr15_-_73295074 | 0.42 |
ENSMUST00000226893.2
|
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr10_+_36383008 | 0.40 |
ENSMUST00000168572.8
|
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr2_-_144174066 | 0.38 |
ENSMUST00000037423.4
|
Ovol2
|
ovo like zinc finger 2 |
| chr3_+_105359641 | 0.33 |
ENSMUST00000098761.10
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3 |
| chr17_-_81035453 | 0.32 |
ENSMUST00000234133.2
ENSMUST00000112389.9 ENSMUST00000025089.9 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr11_-_62348599 | 0.30 |
ENSMUST00000127471.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr17_-_25652750 | 0.28 |
ENSMUST00000159610.8
ENSMUST00000159048.8 ENSMUST00000078496.12 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr2_-_52314837 | 0.28 |
ENSMUST00000036541.8
|
Arl5a
|
ADP-ribosylation factor-like 5A |
| chr8_+_13485451 | 0.27 |
ENSMUST00000167505.3
ENSMUST00000167071.9 |
Tmem255b
|
transmembrane protein 255B |
| chr2_-_157121440 | 0.27 |
ENSMUST00000143663.2
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr6_+_134012916 | 0.26 |
ENSMUST00000164648.2
|
Etv6
|
ets variant 6 |
| chr2_+_153684901 | 0.25 |
ENSMUST00000175856.3
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr16_+_8647959 | 0.24 |
ENSMUST00000023150.7
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chrX_+_13147209 | 0.24 |
ENSMUST00000000804.7
|
Ddx3x
|
DEAD box helicase 3, X-linked |
| chr7_+_19093665 | 0.24 |
ENSMUST00000140836.8
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr5_-_99184894 | 0.22 |
ENSMUST00000031277.7
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr11_-_62348115 | 0.22 |
ENSMUST00000069456.11
ENSMUST00000018645.13 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr2_-_57014015 | 0.21 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr5_-_31350449 | 0.21 |
ENSMUST00000166769.8
|
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chrX_-_47123719 | 0.20 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr4_+_127062924 | 0.17 |
ENSMUST00000046659.14
|
Dlgap3
|
DLG associated protein 3 |
| chr15_-_73294923 | 0.17 |
ENSMUST00000226848.2
ENSMUST00000226466.2 ENSMUST00000226988.3 |
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr4_-_148215135 | 0.14 |
ENSMUST00000030862.5
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr5_-_135518098 | 0.14 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr5_-_99185201 | 0.12 |
ENSMUST00000161490.8
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr13_+_94495457 | 0.12 |
ENSMUST00000022196.5
|
Ap3b1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr2_-_131194754 | 0.11 |
ENSMUST00000059372.11
|
Rnf24
|
ring finger protein 24 |
| chr3_+_86131970 | 0.05 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr18_+_7869066 | 0.03 |
ENSMUST00000171486.8
ENSMUST00000170932.8 ENSMUST00000167020.8 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr11_+_19874354 | 0.02 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr15_-_73295029 | 0.01 |
ENSMUST00000239146.2
ENSMUST00000110036.11 ENSMUST00000228180.3 |
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr11_+_19874403 | 0.01 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr11_-_88609048 | 0.00 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.4 | 2.2 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 2.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 5.0 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 1.6 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 1.9 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.2 | 1.7 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.2 | 3.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 1.4 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 3.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.6 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 1.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.6 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 1.1 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 1.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 1.4 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 5.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 1.9 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 1.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.5 | 2.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 2.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.4 | 1.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.3 | 2.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 2.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 2.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 1.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 1.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 1.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 3.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 2.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 5.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 3.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 3.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |