Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr1i2
Z-value: 1.99
Transcription factors associated with Nr1i2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1i2
|
ENSMUSG00000022809.5 | Nr1i2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1i2 | mm39_v1_chr16_-_38115172_38115200 | -0.40 | 1.5e-02 | Click! |
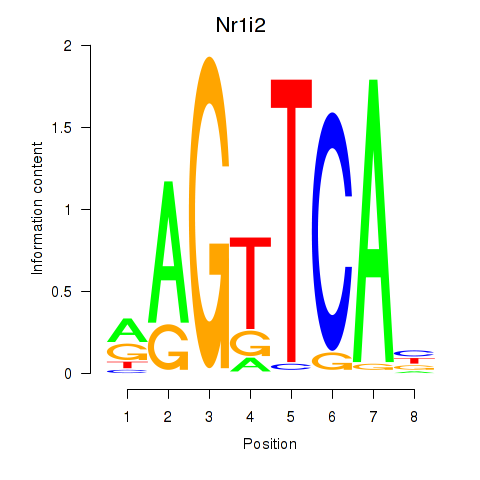
Activity profile of Nr1i2 motif
Sorted Z-values of Nr1i2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1i2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 21.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.2 | 3.7 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.9 | 5.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.8 | 3.9 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.7 | 2.9 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.7 | 4.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 2.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.6 | 4.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.6 | 2.9 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.5 | 2.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.5 | 6.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 1.5 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.5 | 1.5 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) seminal vesicle development(GO:0061107) |
| 0.5 | 2.5 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.5 | 1.4 | GO:0061723 | glycophagy(GO:0061723) |
| 0.4 | 1.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 1.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.4 | 2.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.4 | 1.3 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.4 | 2.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 1.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 1.6 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.3 | 2.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 0.9 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 0.8 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.3 | 8.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.3 | 1.0 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.2 | 2.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 1.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 1.6 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 0.7 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.2 | 1.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 0.8 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.6 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 | 1.3 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 0.9 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.2 | 1.4 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.2 | 0.5 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.2 | 0.7 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 1.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 1.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.2 | 0.5 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.2 | 0.5 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.9 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.6 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 1.5 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.1 | 0.7 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 | 0.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.8 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.1 | 0.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.5 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.5 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 1.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.3 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.1 | 2.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:0002760 | positive regulation of antimicrobial humoral response(GO:0002760) |
| 0.1 | 0.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.5 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.1 | 0.7 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 0.3 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.3 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.1 | 0.3 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.1 | 0.3 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.5 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.6 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 0.4 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.1 | 1.0 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.4 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.4 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 1.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.2 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.1 | 0.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.5 | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.1 | 0.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.2 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 1.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.7 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 0.6 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.1 | 0.2 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.2 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.2 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 0.2 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 | 0.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 1.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.3 | GO:0015867 | ATP transport(GO:0015867) sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 2.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.6 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.0 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.5 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.4 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 1.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0060800 | astrocyte fate commitment(GO:0060018) regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 1.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.4 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.3 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.0 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 1.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.4 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.5 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.6 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.3 | GO:2000344 | cortisol biosynthetic process(GO:0034651) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.6 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 1.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.4 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 2.0 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 1.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.4 | GO:0046851 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.0 | 0.1 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.1 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 1.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.0 | 0.3 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.6 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 | 0.1 | GO:1900738 | psychomotor behavior(GO:0036343) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.0 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 2.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:0044334 | maintenance of DNA repeat elements(GO:0043570) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.0 | 2.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.5 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.2 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 2.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 1.1 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.4 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.6 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.3 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.3 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.5 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.4 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0051642 | centrosome localization(GO:0051642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 2.9 | GO:0097413 | Lewy body(GO:0097413) |
| 0.4 | 1.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 1.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 0.7 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 1.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 0.7 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.2 | 2.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 1.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 0.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.3 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.2 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 2.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 1.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 5.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 1.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 5.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 1.4 | 5.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.0 | 2.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.7 | 4.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.5 | 1.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.5 | 1.6 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.5 | 6.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 1.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.4 | 2.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.4 | 1.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 1.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 1.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 2.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.3 | 0.9 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.3 | 1.1 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.3 | 0.8 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.3 | 1.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.3 | 0.8 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 0.8 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.2 | 17.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.2 | 0.6 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 1.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 0.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.9 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.7 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.1 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.5 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 3.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 4.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.4 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.1 | 3.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 2.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 2.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.2 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 1.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.5 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 1.0 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) |
| 0.0 | 2.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 1.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.4 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.5 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 3.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0005372 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 6.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 4.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 5.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 3.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 3.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 2.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 4.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |