Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr2f6
Z-value: 2.32
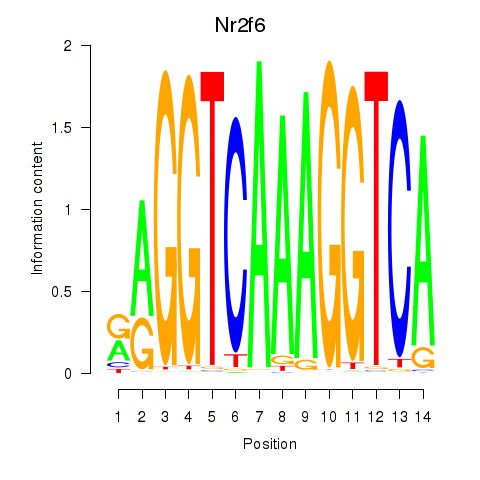
Transcription factors associated with Nr2f6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f6
|
ENSMUSG00000002393.15 | Nr2f6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f6 | mm39_v1_chr8_-_71834543_71834621 | -0.73 | 4.6e-07 | Click! |
Activity profile of Nr2f6 motif
Sorted Z-values of Nr2f6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_9979033 | 12.95 |
ENSMUST00000121418.8
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr10_-_79624758 | 10.57 |
ENSMUST00000020573.13
|
Prss57
|
protease, serine 57 |
| chr2_+_162916551 | 9.64 |
ENSMUST00000142729.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr11_+_116089678 | 9.58 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr10_-_93425553 | 8.27 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr8_+_117822593 | 7.54 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr8_+_71921824 | 7.38 |
ENSMUST00000124745.8
ENSMUST00000138892.2 ENSMUST00000147642.2 |
Dda1
|
DET1 and DDB1 associated 1 |
| chr3_-_89325594 | 7.37 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr9_+_110173253 | 7.22 |
ENSMUST00000199709.3
|
Scap
|
SREBF chaperone |
| chr13_-_59917569 | 6.93 |
ENSMUST00000057115.7
|
Isca1
|
iron-sulfur cluster assembly 1 |
| chr11_+_4936824 | 6.91 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr7_+_98143429 | 6.80 |
ENSMUST00000165205.2
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr16_+_33614378 | 6.74 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr19_+_10019023 | 6.54 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr16_+_33614715 | 6.46 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr2_+_24226857 | 6.26 |
ENSMUST00000114487.9
ENSMUST00000142093.7 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr1_+_63216281 | 5.96 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr13_-_24945844 | 5.79 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chr2_+_90912710 | 5.68 |
ENSMUST00000169852.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr2_+_164647002 | 5.66 |
ENSMUST00000052107.5
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr10_-_35587888 | 5.55 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr17_+_47983587 | 5.45 |
ENSMUST00000152724.2
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr6_+_125122172 | 5.35 |
ENSMUST00000119527.8
ENSMUST00000117675.8 |
Iffo1
|
intermediate filament family orphan 1 |
| chr1_+_63215976 | 5.31 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr5_+_135216090 | 5.09 |
ENSMUST00000002825.6
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr1_-_120197979 | 5.07 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr11_+_101333238 | 4.79 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chrX_-_72954835 | 4.73 |
ENSMUST00000114404.8
ENSMUST00000114407.9 ENSMUST00000114406.9 ENSMUST00000064376.13 ENSMUST00000114405.8 |
Arhgap4
|
Rho GTPase activating protein 4 |
| chr5_+_76988444 | 4.59 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr17_+_48571298 | 4.34 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr11_+_101333115 | 4.14 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr9_+_118335294 | 4.05 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr6_+_85408953 | 4.01 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr5_+_31855009 | 3.97 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr15_-_100320926 | 3.92 |
ENSMUST00000023774.12
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr6_+_136509922 | 3.85 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_+_171216480 | 3.78 |
ENSMUST00000056449.9
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr2_+_75662511 | 3.74 |
ENSMUST00000047232.14
ENSMUST00000111952.9 |
Agps
|
alkylglycerone phosphate synthase |
| chr5_+_31855304 | 3.59 |
ENSMUST00000114515.9
|
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr7_+_26958150 | 3.55 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr7_-_19684654 | 3.49 |
ENSMUST00000043440.8
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr14_-_70414236 | 3.41 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr5_+_31855394 | 3.39 |
ENSMUST00000063813.11
ENSMUST00000071531.12 ENSMUST00000131995.7 ENSMUST00000114507.10 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr7_-_97768725 | 3.38 |
ENSMUST00000107128.8
|
Myo7a
|
myosin VIIA |
| chr2_-_164585102 | 3.31 |
ENSMUST00000103096.10
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr7_-_97768689 | 3.30 |
ENSMUST00000138627.2
ENSMUST00000107127.8 |
Myo7a
|
myosin VIIA |
| chr19_-_61215743 | 3.26 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr4_-_130068484 | 3.12 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr8_-_106434565 | 3.10 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr18_+_63841756 | 3.07 |
ENSMUST00000072726.7
ENSMUST00000235648.2 ENSMUST00000236879.2 |
Wdr7
|
WD repeat domain 7 |
| chr1_-_120198804 | 3.01 |
ENSMUST00000112641.8
|
Steap3
|
STEAP family member 3 |
| chr10_-_22607817 | 3.00 |
ENSMUST00000095794.4
|
Tbpl1
|
TATA box binding protein-like 1 |
| chr7_-_28297565 | 3.00 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr12_-_55126882 | 2.99 |
ENSMUST00000021406.6
|
2700097O09Rik
|
RIKEN cDNA 2700097O09 gene |
| chr1_+_74448535 | 2.98 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr2_+_155453103 | 2.97 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr2_+_75662806 | 2.95 |
ENSMUST00000175646.2
|
Agps
|
alkylglycerone phosphate synthase |
| chr11_+_58269862 | 2.95 |
ENSMUST00000013787.11
ENSMUST00000108826.3 |
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr12_-_57244096 | 2.94 |
ENSMUST00000044634.12
|
Slc25a21
|
solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 |
| chr2_-_157870341 | 2.87 |
ENSMUST00000029179.11
|
Tti1
|
TELO2 interacting protein 1 |
| chr3_-_95041246 | 2.79 |
ENSMUST00000172572.9
ENSMUST00000173462.3 |
Scnm1
|
sodium channel modifier 1 |
| chr6_-_116693849 | 2.74 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr5_-_31854942 | 2.67 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr10_-_13199971 | 2.61 |
ENSMUST00000105543.9
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr2_+_157870606 | 2.54 |
ENSMUST00000109518.8
ENSMUST00000029180.14 |
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr9_+_106339069 | 2.50 |
ENSMUST00000188396.2
|
Pcbp4
|
poly(rC) binding protein 4 |
| chr11_-_83483807 | 2.44 |
ENSMUST00000019071.4
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr10_+_128139191 | 2.43 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr2_+_158636727 | 2.39 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr10_+_128139227 | 2.35 |
ENSMUST00000218315.2
ENSMUST00000219721.2 |
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr10_-_79938487 | 2.34 |
ENSMUST00000042771.8
|
Sbno2
|
strawberry notch 2 |
| chr2_+_157870399 | 2.32 |
ENSMUST00000103123.10
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr15_-_102097387 | 2.27 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr1_+_74545203 | 2.24 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr5_+_145063568 | 2.23 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr10_-_14581203 | 2.21 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr10_-_119075910 | 2.17 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr12_+_55276953 | 2.16 |
ENSMUST00000218879.2
|
Srp54c
|
signal recognition particle 54C |
| chr19_+_47217279 | 2.14 |
ENSMUST00000111807.5
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr13_+_73476629 | 2.11 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr4_-_117146624 | 2.09 |
ENSMUST00000221654.2
|
Rnf220
|
ring finger protein 220 |
| chr2_+_180351910 | 2.08 |
ENSMUST00000029090.9
|
Gid8
|
GID complex subunit 8 |
| chr2_+_153187729 | 2.07 |
ENSMUST00000227428.2
ENSMUST00000109790.2 |
Asxl1
|
ASXL transcriptional regulator 1 |
| chr16_+_43960183 | 2.03 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr11_-_75330415 | 1.90 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr18_-_70605538 | 1.84 |
ENSMUST00000067556.4
|
4930503L19Rik
|
RIKEN cDNA 4930503L19 gene |
| chr11_-_75330302 | 1.82 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr13_+_112425113 | 1.81 |
ENSMUST00000165593.9
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr16_-_18066591 | 1.79 |
ENSMUST00000115645.10
|
Ranbp1
|
RAN binding protein 1 |
| chr1_-_74544946 | 1.77 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr5_+_114141894 | 1.63 |
ENSMUST00000086599.11
|
Dao
|
D-amino acid oxidase |
| chr9_+_108337726 | 1.61 |
ENSMUST00000061209.7
ENSMUST00000193269.2 |
Ccdc71
|
coiled-coil domain containing 71 |
| chr5_-_137739863 | 1.60 |
ENSMUST00000061789.14
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr5_-_110927803 | 1.58 |
ENSMUST00000112426.8
|
Pus1
|
pseudouridine synthase 1 |
| chr3_+_95071617 | 1.58 |
ENSMUST00000168321.8
ENSMUST00000107217.6 ENSMUST00000202315.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr15_-_78377926 | 1.55 |
ENSMUST00000163494.3
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr5_-_110928436 | 1.49 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr11_+_115366470 | 1.48 |
ENSMUST00000035240.7
|
Armc7
|
armadillo repeat containing 7 |
| chr10_+_128583734 | 1.48 |
ENSMUST00000163377.10
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr17_-_35351026 | 1.47 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr12_+_72583114 | 1.44 |
ENSMUST00000044352.8
|
Pcnx4
|
pecanex homolog 4 |
| chr4_-_41697040 | 1.44 |
ENSMUST00000102962.10
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr15_-_81756076 | 1.41 |
ENSMUST00000023117.10
|
Phf5a
|
PHD finger protein 5A |
| chr4_-_136776006 | 1.40 |
ENSMUST00000049583.8
|
Zbtb40
|
zinc finger and BTB domain containing 40 |
| chr19_-_47525437 | 1.38 |
ENSMUST00000182808.8
ENSMUST00000182714.2 ENSMUST00000049369.16 |
Stn1
|
STN1, CST complex subunit |
| chr11_-_33463722 | 1.38 |
ENSMUST00000102815.10
|
Ranbp17
|
RAN binding protein 17 |
| chr6_+_124690060 | 1.38 |
ENSMUST00000130279.2
|
Phb2
|
prohibitin 2 |
| chr4_+_132291369 | 1.33 |
ENSMUST00000070690.8
|
Ptafr
|
platelet-activating factor receptor |
| chr13_+_112425049 | 1.29 |
ENSMUST00000168684.2
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr3_+_115681788 | 1.27 |
ENSMUST00000196804.5
ENSMUST00000106505.8 ENSMUST00000043342.10 |
Dph5
|
diphthamide biosynthesis 5 |
| chr4_+_110254907 | 1.25 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr3_+_115681486 | 1.23 |
ENSMUST00000189799.7
ENSMUST00000200258.2 |
Dph5
|
diphthamide biosynthesis 5 |
| chr7_+_82297803 | 1.22 |
ENSMUST00000141726.8
ENSMUST00000179489.8 ENSMUST00000039881.4 |
Efl1
|
elongation factor like GTPase 1 |
| chr9_-_22046970 | 1.18 |
ENSMUST00000165735.9
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr11_-_99134885 | 1.14 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr1_-_74639723 | 1.14 |
ENSMUST00000127938.8
ENSMUST00000154874.8 |
Rnf25
|
ring finger protein 25 |
| chr9_+_20914211 | 1.13 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr7_-_19449319 | 1.11 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr5_-_134205559 | 1.07 |
ENSMUST00000076228.3
|
Rcc1l
|
reculator of chromosome condensation 1 like |
| chr4_-_117539431 | 1.05 |
ENSMUST00000102687.4
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr1_+_180553569 | 1.05 |
ENSMUST00000027780.6
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr5_+_145282064 | 1.05 |
ENSMUST00000079268.9
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr5_+_114142842 | 1.05 |
ENSMUST00000161610.6
|
Dao
|
D-amino acid oxidase |
| chr19_-_45986919 | 1.03 |
ENSMUST00000045396.9
|
Armh3
|
armadillo-like helical domain containing 3 |
| chr4_+_110254858 | 1.00 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr9_-_109575157 | 0.98 |
ENSMUST00000071917.4
|
Fbxw26
|
F-box and WD-40 domain protein 26 |
| chr1_+_135727571 | 0.98 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr9_+_104424466 | 0.92 |
ENSMUST00000098443.9
|
Cpne4
|
copine IV |
| chr1_-_65158717 | 0.92 |
ENSMUST00000144760.3
|
Gm28845
|
predicted gene 28845 |
| chr14_+_60615128 | 0.91 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr15_+_86098660 | 0.90 |
ENSMUST00000063414.9
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr8_+_66964401 | 0.87 |
ENSMUST00000002025.5
ENSMUST00000183187.2 |
Tktl2
|
transketolase-like 2 |
| chr16_-_4442802 | 0.87 |
ENSMUST00000014445.7
|
Pam16
|
presequence translocase-asssociated motor 16 homolog (S. cerevisiae) |
| chr9_+_20914012 | 0.86 |
ENSMUST00000003386.7
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr15_-_102097466 | 0.85 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr9_-_108338111 | 0.77 |
ENSMUST00000193895.6
|
Klhdc8b
|
kelch domain containing 8B |
| chrX_+_100492684 | 0.75 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr2_+_25132941 | 0.71 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr12_-_104718159 | 0.70 |
ENSMUST00000041987.7
|
Dicer1
|
dicer 1, ribonuclease type III |
| chr10_+_78410803 | 0.69 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr1_-_93088562 | 0.69 |
ENSMUST00000143419.2
|
Mab21l4
|
mab-21-like 4 |
| chr11_-_84761637 | 0.64 |
ENSMUST00000168434.8
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr19_-_9978987 | 0.61 |
ENSMUST00000117346.2
|
Best1
|
bestrophin 1 |
| chr12_+_55201889 | 0.59 |
ENSMUST00000110708.4
|
Srp54b
|
signal recognition particle 54B |
| chr6_+_83133441 | 0.59 |
ENSMUST00000203203.2
|
1700003E16Rik
|
RIKEN cDNA 1700003E16 gene |
| chr17_+_35069347 | 0.58 |
ENSMUST00000097343.11
ENSMUST00000173357.8 ENSMUST00000173065.8 ENSMUST00000165953.3 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr3_+_95041399 | 0.57 |
ENSMUST00000066386.6
|
Lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr11_-_84761538 | 0.54 |
ENSMUST00000170741.2
ENSMUST00000172405.8 ENSMUST00000100686.10 ENSMUST00000108081.9 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr17_-_23805187 | 0.49 |
ENSMUST00000227952.2
ENSMUST00000115516.11 |
Zfp13
|
zinc finger protein 13 |
| chr5_-_31359559 | 0.48 |
ENSMUST00000202929.2
ENSMUST00000201231.2 ENSMUST00000114590.8 |
Zfp513
|
zinc finger protein 513 |
| chr13_+_22574543 | 0.45 |
ENSMUST00000226157.2
ENSMUST00000227326.2 ENSMUST00000228726.2 |
Vmn1r200
|
vomeronasal 1 receptor 200 |
| chr7_+_30758767 | 0.45 |
ENSMUST00000039775.9
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr1_-_74323536 | 0.44 |
ENSMUST00000190008.2
|
Aamp
|
angio-associated migratory protein |
| chr2_-_164646794 | 0.43 |
ENSMUST00000103094.11
ENSMUST00000017451.7 |
Acot8
|
acyl-CoA thioesterase 8 |
| chr6_+_83985684 | 0.42 |
ENSMUST00000203803.3
ENSMUST00000204591.3 ENSMUST00000113823.8 ENSMUST00000153860.4 |
Dysf
|
dysferlin |
| chr9_-_110818679 | 0.40 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr15_+_89417017 | 0.40 |
ENSMUST00000167173.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr11_-_97242842 | 0.38 |
ENSMUST00000093942.5
|
Gpr179
|
G protein-coupled receptor 179 |
| chr14_-_52277310 | 0.33 |
ENSMUST00000216907.2
ENSMUST00000214071.2 ENSMUST00000216188.2 |
Olfr221
|
olfactory receptor 221 |
| chr16_+_18066730 | 0.30 |
ENSMUST00000115640.8
ENSMUST00000140206.8 |
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr6_+_21986445 | 0.28 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_+_58035640 | 0.28 |
ENSMUST00000111269.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr4_-_94538329 | 0.28 |
ENSMUST00000107101.2
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr14_+_58036071 | 0.28 |
ENSMUST00000111268.8
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr7_+_140239843 | 0.27 |
ENSMUST00000218865.2
|
Olfr539
|
olfactory receptor 539 |
| chr2_-_32665342 | 0.26 |
ENSMUST00000161089.8
ENSMUST00000066478.9 ENSMUST00000161950.8 ENSMUST00000091059.12 |
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr12_+_55126999 | 0.26 |
ENSMUST00000220578.2
ENSMUST00000221655.2 |
Srp54a
|
signal recognition particle 54A |
| chr4_+_152160713 | 0.26 |
ENSMUST00000239025.2
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr11_-_69791712 | 0.25 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr17_+_28910302 | 0.24 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr7_-_18390645 | 0.23 |
ENSMUST00000094793.12
ENSMUST00000182128.2 |
Psg21
|
pregnancy-specific glycoprotein 21 |
| chr17_-_35069136 | 0.22 |
ENSMUST00000046022.16
|
Skiv2l
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr6_+_83985495 | 0.22 |
ENSMUST00000113821.8
|
Dysf
|
dysferlin |
| chr11_-_69791756 | 0.21 |
ENSMUST00000018714.13
ENSMUST00000128046.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr14_+_54032814 | 0.21 |
ENSMUST00000103671.4
|
Trav13-5
|
T cell receptor alpha variable 13-5 |
| chr6_+_114380627 | 0.20 |
ENSMUST00000161650.3
|
Hrh1
|
histamine receptor H1 |
| chr6_+_114380786 | 0.16 |
ENSMUST00000161220.2
|
Hrh1
|
histamine receptor H1 |
| chr1_+_57445487 | 0.16 |
ENSMUST00000027114.6
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr6_+_124689241 | 0.15 |
ENSMUST00000004375.16
|
Phb2
|
prohibitin 2 |
| chr6_+_21985902 | 0.14 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_+_149942140 | 0.13 |
ENSMUST00000065745.10
ENSMUST00000110496.5 ENSMUST00000201612.2 |
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr7_-_89590334 | 0.13 |
ENSMUST00000207309.2
ENSMUST00000130609.3 |
Hikeshi
|
heat shock protein nuclear import factor |
| chr17_-_36220924 | 0.12 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr15_+_77583104 | 0.11 |
ENSMUST00000096358.6
|
Apol7e
|
apolipoprotein L 7e |
| chr7_-_43906629 | 0.10 |
ENSMUST00000012921.9
|
Acp4
|
acid phosphatase 4 |
| chr19_-_27407206 | 0.09 |
ENSMUST00000076219.6
|
Pum3
|
pumilio RNA-binding family member 3 |
| chr1_+_91072778 | 0.09 |
ENSMUST00000188818.2
ENSMUST00000094698.2 |
Rbm44
|
RNA binding motif protein 44 |
| chr16_+_18066825 | 0.09 |
ENSMUST00000100099.10
|
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr13_-_73476561 | 0.08 |
ENSMUST00000222930.2
ENSMUST00000223293.2 ENSMUST00000022097.6 |
Ndufs6
|
NADH:ubiquinone oxidoreductase core subunit S6 |
| chr6_-_137626207 | 0.08 |
ENSMUST00000134630.6
ENSMUST00000058210.13 ENSMUST00000111878.8 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr6_+_113214804 | 0.07 |
ENSMUST00000113146.9
|
Mtmr14
|
myotubularin related protein 14 |
| chr13_-_64396422 | 0.07 |
ENSMUST00000109769.10
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr7_-_89590230 | 0.06 |
ENSMUST00000075010.12
|
Hikeshi
|
heat shock protein nuclear import factor |
| chr17_-_36220518 | 0.03 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr17_+_49922129 | 0.02 |
ENSMUST00000162854.2
|
Kif6
|
kinesin family member 6 |
| chr15_-_77331660 | 0.01 |
ENSMUST00000089469.7
|
Apol7b
|
apolipoprotein L 7b |
| chr2_-_32665637 | 0.01 |
ENSMUST00000161958.2
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr11_-_69791774 | 0.01 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr15_-_77330396 | 0.01 |
ENSMUST00000229434.2
|
Apol7b
|
apolipoprotein L 7b |
| chr1_-_74323795 | 0.00 |
ENSMUST00000178235.8
ENSMUST00000006462.14 |
Aamp
|
angio-associated migratory protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.6 | 8.1 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 1.3 | 6.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.1 | 5.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.1 | 5.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.0 | 3.0 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.9 | 2.7 | GO:0046436 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.9 | 6.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.8 | 13.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.7 | 3.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.7 | 2.7 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.6 | 3.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.6 | 3.0 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.6 | 2.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.6 | 6.7 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.5 | 4.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.5 | 2.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.5 | 3.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.5 | 2.5 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.5 | 2.5 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.5 | 1.5 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.5 | 3.7 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.4 | 2.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.4 | 1.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.4 | 6.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.4 | 11.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.4 | 6.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.4 | 4.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.4 | 4.7 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.4 | 2.2 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.4 | 1.4 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 1.3 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.3 | 2.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 1.8 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.3 | 7.2 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.3 | 5.0 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.3 | 3.6 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.2 | 1.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 5.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 2.5 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 2.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 6.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.2 | 12.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 11.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 8.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 6.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.7 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 1.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.2 | 3.3 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 1.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 9.6 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 7.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.5 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 1.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 5.8 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 4.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.4 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 1.0 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 2.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 3.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 2.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 8.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 2.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 7.4 | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032434) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 2.9 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 1.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 1.5 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:1990879 | CST complex(GO:1990879) |
| 3.5 | 10.6 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.6 | 11.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.6 | 9.6 | GO:0031523 | Myb complex(GO:0031523) |
| 1.4 | 8.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.1 | 10.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 1.0 | 4.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.8 | 6.7 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.6 | 2.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.5 | 3.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.4 | 3.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.3 | 1.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 1.4 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 2.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 2.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 2.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 8.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 3.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 6.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 5.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 3.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 8.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 7.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 4.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.1 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 3.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 3.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 11.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 4.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 2.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 5.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.9 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.2 | 7.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.1 | 3.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.0 | 8.1 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.8 | 9.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.8 | 5.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.7 | 2.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.5 | 2.1 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.5 | 1.5 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.5 | 12.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 3.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.4 | 5.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 3.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 1.8 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.3 | 0.9 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.3 | 6.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 12.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 11.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 1.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 10.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.2 | 4.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 5.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 6.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 3.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 5.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 3.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.4 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 1.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.7 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 4.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.4 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 7.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 6.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 2.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 13.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.9 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 2.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 5.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 6.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 4.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 7.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 7.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 2.6 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 6.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.5 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 3.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 6.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 3.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 3.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 7.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 11.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 6.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 7.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 3.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 8.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 14.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 10.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 5.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.6 | 13.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.5 | 6.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 5.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.3 | 9.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 7.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 5.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 6.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 20.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 3.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 4.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.8 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 4.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |