Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr3c2
Z-value: 0.38
Transcription factors associated with Nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr3c2
|
ENSMUSG00000031618.14 | Nr3c2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c2 | mm39_v1_chr8_+_77626400_77626456 | -0.37 | 2.8e-02 | Click! |
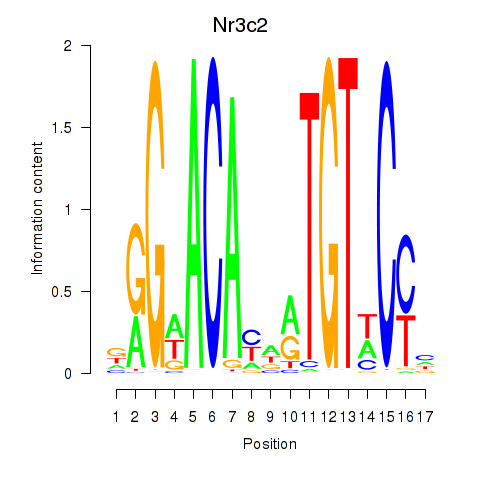
Activity profile of Nr3c2 motif
Sorted Z-values of Nr3c2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr3c2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94905710 | 2.34 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr8_+_71858647 | 0.89 |
ENSMUST00000119976.8
ENSMUST00000120725.2 |
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr4_-_149221998 | 0.84 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr10_-_117118226 | 0.76 |
ENSMUST00000092163.9
|
Lyz2
|
lysozyme 2 |
| chr11_+_68989763 | 0.72 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr4_-_149222057 | 0.70 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr9_+_123195986 | 0.63 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr16_+_38167352 | 0.55 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr5_+_31855009 | 0.49 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr5_-_31854942 | 0.45 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr15_-_81931783 | 0.36 |
ENSMUST00000080622.9
|
Snu13
|
SNU13 homolog, small nuclear ribonucleoprotein (U4/U6.U5) |
| chr8_-_25730878 | 0.34 |
ENSMUST00000210488.2
ENSMUST00000210933.2 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr11_+_96822213 | 0.33 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr5_+_123280250 | 0.30 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr16_+_16844217 | 0.27 |
ENSMUST00000232067.2
|
Mapk1
|
mitogen-activated protein kinase 1 |
| chr7_-_141934524 | 0.24 |
ENSMUST00000209263.2
|
Gm49369
|
predicted gene, 49369 |
| chr11_-_75345482 | 0.23 |
ENSMUST00000173320.8
|
Wdr81
|
WD repeat domain 81 |
| chr4_-_9643636 | 0.16 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr6_-_69220672 | 0.16 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr6_+_136495784 | 0.15 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_-_135494775 | 0.13 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr10_-_14593935 | 0.13 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr6_-_69584812 | 0.12 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr6_+_136495818 | 0.11 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr15_-_85705935 | 0.10 |
ENSMUST00000064370.6
|
Pkdrej
|
polycystin (PKD) family receptor for egg jelly |
| chr6_+_52691204 | 0.09 |
ENSMUST00000138040.8
ENSMUST00000129660.2 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr6_-_69020489 | 0.09 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr6_+_34840151 | 0.08 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr11_-_71092282 | 0.07 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr1_-_74974707 | 0.07 |
ENSMUST00000094844.4
|
Cfap65
|
cilia and flagella associated protein 65 |
| chr11_-_71092124 | 0.07 |
ENSMUST00000108514.10
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr9_-_75504926 | 0.07 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr6_-_69204417 | 0.07 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr6_-_69377328 | 0.07 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr11_+_50905063 | 0.06 |
ENSMUST00000217480.2
ENSMUST00000215409.2 |
Olfr54
|
olfactory receptor 54 |
| chr6_-_69037208 | 0.06 |
ENSMUST00000103343.4
|
Igkv4-78
|
immunoglobulin kappa variable 4-78 |
| chr17_+_23562715 | 0.06 |
ENSMUST00000168175.3
ENSMUST00000234796.2 |
Vmn2r115
|
vomeronasal 2, receptor 115 |
| chr8_-_95328348 | 0.05 |
ENSMUST00000212547.2
ENSMUST00000212507.2 ENSMUST00000034226.8 |
Psme3ip1
|
proteasome activator subunit 3 interacting protein 1 |
| chr14_+_35816874 | 0.05 |
ENSMUST00000226305.2
|
4930474N05Rik
|
RIKEN cDNA 4930474N05 gene |
| chr6_-_69609162 | 0.05 |
ENSMUST00000199437.2
|
Igkv4-54
|
immunoglobulin kappa chain variable 4-54 |
| chr5_+_112436599 | 0.05 |
ENSMUST00000151947.3
|
Tpst2
|
protein-tyrosine sulfotransferase 2 |
| chr1_+_173959083 | 0.05 |
ENSMUST00000214751.2
|
Olfr424
|
olfactory receptor 424 |
| chr17_-_22792463 | 0.05 |
ENSMUST00000092491.7
ENSMUST00000234223.2 ENSMUST00000234296.2 ENSMUST00000234027.2 |
Vmn2r111
|
vomeronasal 2, receptor 111 |
| chr14_+_54000594 | 0.04 |
ENSMUST00000103589.6
|
Trav14-3
|
T cell receptor alpha variable 14-3 |
| chr10_+_26698556 | 0.03 |
ENSMUST00000135866.2
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr10_+_128061699 | 0.03 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr7_+_106630381 | 0.03 |
ENSMUST00000213623.2
|
Olfr713
|
olfactory receptor 713 |
| chr11_+_98277276 | 0.03 |
ENSMUST00000041301.8
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr9_+_99494550 | 0.03 |
ENSMUST00000042553.8
|
A4gnt
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr7_+_118199375 | 0.02 |
ENSMUST00000121744.9
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr6_-_6882068 | 0.02 |
ENSMUST00000142635.2
ENSMUST00000052609.9 |
Dlx5
|
distal-less homeobox 5 |
| chr17_+_22819932 | 0.02 |
ENSMUST00000097381.5
ENSMUST00000234882.2 |
Vmn2r112
|
vomeronasal 2, receptor 112 |
| chr17_-_23531402 | 0.01 |
ENSMUST00000168033.3
|
Vmn2r114
|
vomeronasal 2, receptor 114 |
| chr13_-_120374288 | 0.01 |
ENSMUST00000179502.2
|
Gm21761
|
predicted gene, 21761 |
| chr13_+_120616163 | 0.01 |
ENSMUST00000179071.2
|
Gm20767
|
predicted gene, 20767 |
| chr6_-_126512375 | 0.01 |
ENSMUST00000060972.5
|
Kcna5
|
potassium voltage-gated channel, shaker-related subfamily, member 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.3 | 2.3 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 0.6 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.9 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.7 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 1.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.9 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0031428 | dense fibrillar component(GO:0001651) box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.4 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 2.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 1.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |