Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr5a2
Z-value: 2.62
Transcription factors associated with Nr5a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr5a2
|
ENSMUSG00000026398.15 | Nr5a2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr5a2 | mm39_v1_chr1_-_136888118_136888234 | -0.69 | 3.2e-06 | Click! |
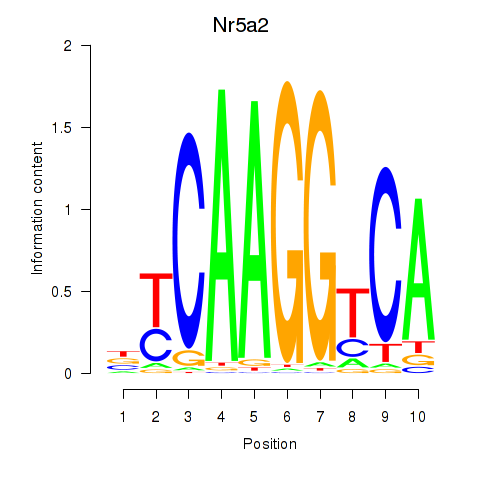
Activity profile of Nr5a2 motif
Sorted Z-values of Nr5a2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr5a2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90638580 | 36.93 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chrX_-_7834057 | 19.63 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr3_+_14951264 | 19.03 |
ENSMUST00000192609.6
|
Car2
|
carbonic anhydrase 2 |
| chr16_+_32427738 | 18.12 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr16_+_32427789 | 16.95 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr11_+_74510413 | 14.72 |
ENSMUST00000100866.3
|
Ccdc92b
|
coiled-coil domain containing 92B |
| chr2_-_131001916 | 14.64 |
ENSMUST00000103188.10
ENSMUST00000133602.8 ENSMUST00000028800.12 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr15_+_89218601 | 14.46 |
ENSMUST00000023282.9
|
Miox
|
myo-inositol oxygenase |
| chr6_-_115739284 | 13.97 |
ENSMUST00000166254.7
ENSMUST00000170625.8 |
Tmem40
|
transmembrane protein 40 |
| chr2_-_28453374 | 13.43 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr2_+_131028861 | 13.00 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr4_-_137157824 | 12.28 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr3_+_14951478 | 12.26 |
ENSMUST00000029078.9
|
Car2
|
carbonic anhydrase 2 |
| chr1_-_167221344 | 11.57 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr7_+_100143250 | 11.32 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr2_-_25911691 | 11.02 |
ENSMUST00000036509.14
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr2_-_25911544 | 10.68 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr7_+_110368037 | 10.61 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr15_-_103161237 | 10.17 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr8_+_95722289 | 10.17 |
ENSMUST00000211984.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_+_65008735 | 10.14 |
ENSMUST00000213533.2
ENSMUST00000035499.5 ENSMUST00000077696.13 ENSMUST00000166273.2 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chrX_+_74425990 | 9.87 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr19_-_11618165 | 9.79 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr8_+_85696695 | 9.68 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr19_-_11618192 | 8.99 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr8_+_85696453 | 8.91 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_85696396 | 8.72 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr6_-_41012435 | 8.10 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr19_+_6450553 | 7.99 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr12_-_110945415 | 7.32 |
ENSMUST00000135131.2
ENSMUST00000043459.13 ENSMUST00000128353.8 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr6_+_90596123 | 7.29 |
ENSMUST00000032177.10
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr17_+_29042640 | 7.06 |
ENSMUST00000233088.2
ENSMUST00000233182.2 ENSMUST00000233520.2 |
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr10_-_80691009 | 6.92 |
ENSMUST00000220225.2
ENSMUST00000035775.9 |
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_103788321 | 6.87 |
ENSMUST00000156813.8
ENSMUST00000170926.8 |
Lmo2
|
LIM domain only 2 |
| chr2_+_32477069 | 6.21 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr5_+_118165808 | 6.06 |
ENSMUST00000031304.14
|
Tesc
|
tescalcin |
| chr6_+_30541581 | 6.03 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr11_-_3454766 | 5.98 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr19_+_6449887 | 5.96 |
ENSMUST00000146601.8
ENSMUST00000150713.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr15_-_63932288 | 5.92 |
ENSMUST00000063838.11
ENSMUST00000228908.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr9_+_44245981 | 5.85 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr10_-_62363217 | 5.82 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chr15_-_79976016 | 5.75 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr17_+_29042544 | 5.71 |
ENSMUST00000140587.9
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr7_+_4743114 | 5.62 |
ENSMUST00000098853.9
ENSMUST00000130215.8 ENSMUST00000108582.10 ENSMUST00000108583.9 |
Kmt5c
|
lysine methyltransferase 5C |
| chr6_+_113508636 | 5.58 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr2_+_129435448 | 5.45 |
ENSMUST00000049262.14
ENSMUST00000163034.8 ENSMUST00000160276.2 |
Sirpa
|
signal-regulatory protein alpha |
| chr11_+_58839716 | 5.45 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chr4_-_116228921 | 5.42 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr7_-_126398165 | 5.40 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr15_-_79169671 | 5.31 |
ENSMUST00000170955.2
ENSMUST00000165408.8 |
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr13_+_21906214 | 5.13 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr13_+_21994588 | 5.10 |
ENSMUST00000091745.6
|
H2ac23
|
H2A clustered histone 23 |
| chrX_-_73293425 | 4.82 |
ENSMUST00000114299.8
|
Flna
|
filamin, alpha |
| chr11_+_96941637 | 4.80 |
ENSMUST00000168565.2
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr11_+_96941420 | 4.74 |
ENSMUST00000090020.13
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr10_+_80100812 | 4.64 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr19_+_6450641 | 4.51 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr12_-_110945052 | 4.47 |
ENSMUST00000140788.8
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr2_+_25152600 | 4.41 |
ENSMUST00000114336.4
|
Tprn
|
taperin |
| chr6_-_76474767 | 4.40 |
ENSMUST00000097218.7
|
Gm9008
|
predicted pseudogene 9008 |
| chr4_-_131802606 | 4.33 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr15_-_98626002 | 4.30 |
ENSMUST00000003445.8
|
Fkbp11
|
FK506 binding protein 11 |
| chr19_-_7218363 | 4.30 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr12_+_102521225 | 4.27 |
ENSMUST00000021610.7
|
Chga
|
chromogranin A |
| chr4_+_131600918 | 4.22 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr3_-_107992662 | 4.21 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr1_+_136059101 | 4.21 |
ENSMUST00000075164.11
|
Kif21b
|
kinesin family member 21B |
| chr12_-_110945376 | 4.18 |
ENSMUST00000142012.2
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr19_+_8568618 | 4.04 |
ENSMUST00000170817.2
ENSMUST00000010251.11 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr3_-_54823287 | 4.02 |
ENSMUST00000070342.4
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr4_+_63477018 | 3.88 |
ENSMUST00000077709.11
|
Tmem268
|
transmembrane protein 268 |
| chr9_+_107879700 | 3.85 |
ENSMUST00000035214.11
ENSMUST00000176854.7 ENSMUST00000175874.2 |
Ip6k1
|
inositol hexaphosphate kinase 1 |
| chr13_-_22227114 | 3.80 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr7_-_33929667 | 3.77 |
ENSMUST00000206415.2
|
Gpi1
|
glucose-6-phosphate isomerase 1 |
| chr1_+_75377616 | 3.76 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr7_+_46496506 | 3.76 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chrX_+_70600481 | 3.76 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr3_-_83947416 | 3.75 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr7_+_130467564 | 3.72 |
ENSMUST00000075181.11
ENSMUST00000151119.9 ENSMUST00000048180.12 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr19_+_6449776 | 3.69 |
ENSMUST00000113468.8
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr15_-_63932176 | 3.67 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr7_+_46496929 | 3.66 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr15_-_64254754 | 3.63 |
ENSMUST00000177374.8
ENSMUST00000110114.10 ENSMUST00000110115.9 ENSMUST00000023008.16 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr2_+_26518456 | 3.63 |
ENSMUST00000074240.4
|
Dipk1b
|
divergent protein kinase domain 1B |
| chr7_-_33929708 | 3.62 |
ENSMUST00000038027.6
|
Gpi1
|
glucose-6-phosphate isomerase 1 |
| chr15_+_78783867 | 3.60 |
ENSMUST00000134703.8
ENSMUST00000061239.14 ENSMUST00000109698.9 |
Gm49510
Sh3bp1
|
predicted gene, 49510 SH3-domain binding protein 1 |
| chr5_+_33493529 | 3.55 |
ENSMUST00000202113.2
|
Maea
|
macrophage erythroblast attacher |
| chr2_-_167334746 | 3.55 |
ENSMUST00000109211.9
ENSMUST00000057627.16 |
Spata2
|
spermatogenesis associated 2 |
| chrX_+_141608694 | 3.51 |
ENSMUST00000112888.2
|
Tmem164
|
transmembrane protein 164 |
| chr5_+_137628377 | 3.49 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr4_+_34893772 | 3.48 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr8_-_106198112 | 3.48 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr7_+_46496552 | 3.48 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr16_+_35590745 | 3.38 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr13_-_21937997 | 3.35 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr13_+_22227359 | 3.34 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr15_-_102425241 | 3.12 |
ENSMUST00000169162.8
ENSMUST00000023812.10 ENSMUST00000165174.8 ENSMUST00000169367.8 ENSMUST00000169377.8 |
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr10_-_80184238 | 3.08 |
ENSMUST00000095446.10
ENSMUST00000105352.2 |
Adamtsl5
|
ADAMTS-like 5 |
| chr7_-_105131407 | 3.05 |
ENSMUST00000047040.4
|
Cavin3
|
caveolae associated 3 |
| chr19_+_47079187 | 3.02 |
ENSMUST00000072141.4
|
Pdcd11
|
programmed cell death 11 |
| chr9_+_107468146 | 3.01 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr12_+_104998895 | 2.99 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr1_-_79838897 | 2.94 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr17_+_29251602 | 2.90 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr12_-_111638722 | 2.79 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr7_-_35096133 | 2.78 |
ENSMUST00000154597.2
ENSMUST00000032704.12 |
Faap24
|
Fanconi anemia core complex associated protein 24 |
| chr13_+_21900554 | 2.75 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr16_-_20245071 | 2.71 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr16_-_20245138 | 2.70 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_+_135727571 | 2.69 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr16_-_8455525 | 2.66 |
ENSMUST00000052505.10
|
Tmem186
|
transmembrane protein 186 |
| chr4_-_126096376 | 2.59 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr8_-_106022676 | 2.58 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chr3_-_86828140 | 2.53 |
ENSMUST00000029719.14
|
Dclk2
|
doublecortin-like kinase 2 |
| chr17_-_24360516 | 2.52 |
ENSMUST00000115411.8
ENSMUST00000115409.9 ENSMUST00000115407.9 ENSMUST00000102927.10 |
Pdpk1
|
3-phosphoinositide dependent protein kinase 1 |
| chr8_-_106553822 | 2.50 |
ENSMUST00000239468.2
ENSMUST00000041400.6 |
Ranbp10
|
RAN binding protein 10 |
| chr2_+_155453103 | 2.46 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr2_-_93283024 | 2.46 |
ENSMUST00000111257.8
ENSMUST00000145553.8 |
Cd82
|
CD82 antigen |
| chr11_+_5008110 | 2.44 |
ENSMUST00000037218.2
|
Rasl10a
|
RAS-like, family 10, member A |
| chr1_-_133537953 | 2.39 |
ENSMUST00000164574.2
ENSMUST00000166291.8 ENSMUST00000164096.2 ENSMUST00000166915.8 |
Snrpe
|
small nuclear ribonucleoprotein E |
| chr7_-_79115915 | 2.32 |
ENSMUST00000073889.14
|
Polg
|
polymerase (DNA directed), gamma |
| chr17_-_27247581 | 2.30 |
ENSMUST00000143158.3
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr5_-_137609634 | 2.30 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr8_-_105350898 | 2.29 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr15_-_77653979 | 2.28 |
ENSMUST00000229259.2
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr8_-_70975813 | 2.27 |
ENSMUST00000121623.8
ENSMUST00000093456.12 ENSMUST00000118850.8 |
Kxd1
|
KxDL motif containing 1 |
| chr5_-_5744024 | 2.26 |
ENSMUST00000115425.9
ENSMUST00000115427.8 ENSMUST00000115424.9 ENSMUST00000015797.11 |
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr11_+_98632631 | 2.23 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr5_+_34527230 | 2.22 |
ENSMUST00000180376.8
|
Fam193a
|
family with sequence homology 193, member A |
| chr11_+_96820091 | 2.20 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr5_-_5744559 | 2.17 |
ENSMUST00000115426.9
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr15_-_64254585 | 2.16 |
ENSMUST00000176384.8
ENSMUST00000175799.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr13_-_115068626 | 2.15 |
ENSMUST00000056117.10
|
Itga2
|
integrin alpha 2 |
| chr15_+_98006346 | 2.15 |
ENSMUST00000051226.8
|
Pfkm
|
phosphofructokinase, muscle |
| chrX_+_156482116 | 2.14 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr11_+_121593582 | 2.14 |
ENSMUST00000125580.2
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr11_+_96820220 | 2.13 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr5_-_137609691 | 2.11 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr15_+_84553801 | 2.10 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr1_-_52230062 | 2.09 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr11_+_31822211 | 2.09 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_-_137137088 | 2.08 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr7_-_100232276 | 2.04 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr5_+_129802127 | 2.03 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr19_-_46315543 | 2.02 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr11_+_3939924 | 1.98 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr2_-_30305472 | 1.96 |
ENSMUST00000134120.2
ENSMUST00000102854.10 |
Crat
|
carnitine acetyltransferase |
| chr9_+_57921954 | 1.95 |
ENSMUST00000034874.14
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr4_+_132857816 | 1.92 |
ENSMUST00000084241.12
ENSMUST00000138831.2 |
Wasf2
|
WASP family, member 2 |
| chr17_-_27842237 | 1.91 |
ENSMUST00000062397.13
ENSMUST00000176876.8 ENSMUST00000146321.3 |
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr19_-_5416339 | 1.91 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr4_-_45108038 | 1.91 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr14_+_20979466 | 1.88 |
ENSMUST00000022369.9
|
Vcl
|
vinculin |
| chr2_-_60793536 | 1.87 |
ENSMUST00000028347.13
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr19_-_60862964 | 1.86 |
ENSMUST00000025961.7
|
Prdx3
|
peroxiredoxin 3 |
| chr1_-_75196496 | 1.83 |
ENSMUST00000186758.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr5_-_5744326 | 1.77 |
ENSMUST00000148333.8
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr7_-_97794679 | 1.73 |
ENSMUST00000098281.4
|
Omp
|
olfactory marker protein |
| chr7_+_127078371 | 1.71 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr19_+_4203603 | 1.70 |
ENSMUST00000236632.2
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr8_-_105350881 | 1.63 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr15_-_99355623 | 1.62 |
ENSMUST00000023747.14
|
Nckap5l
|
NCK-associated protein 5-like |
| chr2_-_174314672 | 1.61 |
ENSMUST00000117442.8
ENSMUST00000141100.2 ENSMUST00000120822.2 |
Prelid3b
|
PRELI domain containing 3B |
| chr7_-_4448631 | 1.58 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr7_-_108774367 | 1.57 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr15_-_98729333 | 1.56 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr10_-_59277570 | 1.56 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr16_+_4704103 | 1.56 |
ENSMUST00000023159.10
ENSMUST00000070658.16 ENSMUST00000229038.2 |
Mgrn1
|
mahogunin, ring finger 1 |
| chr7_-_79115760 | 1.48 |
ENSMUST00000125562.2
|
Polg
|
polymerase (DNA directed), gamma |
| chr12_-_84240781 | 1.48 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr4_-_138858340 | 1.48 |
ENSMUST00000143971.2
|
Micos10
|
mitochondrial contact site and cristae organizing system subunit 10 |
| chr16_-_4607848 | 1.47 |
ENSMUST00000004173.12
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr13_+_23930717 | 1.45 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr3_+_63883527 | 1.45 |
ENSMUST00000029405.8
|
Gmps
|
guanine monophosphate synthetase |
| chr8_-_105350533 | 1.44 |
ENSMUST00000212662.2
|
Cdh16
|
cadherin 16 |
| chr6_+_87864796 | 1.44 |
ENSMUST00000113607.10
ENSMUST00000049966.6 |
Copg1
|
coatomer protein complex, subunit gamma 1 |
| chr19_-_4665668 | 1.44 |
ENSMUST00000113822.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chrX_+_20529137 | 1.43 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr19_-_4665509 | 1.42 |
ENSMUST00000053597.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr7_-_130148984 | 1.42 |
ENSMUST00000160289.9
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr16_-_43800109 | 1.40 |
ENSMUST00000231700.2
|
Zdhhc23
|
zinc finger, DHHC domain containing 23 |
| chr16_+_4704166 | 1.37 |
ENSMUST00000230990.2
|
Mgrn1
|
mahogunin, ring finger 1 |
| chr19_-_5733371 | 1.37 |
ENSMUST00000127876.8
|
Pcnx3
|
pecanex homolog 3 |
| chr4_-_126096551 | 1.36 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr9_+_45311000 | 1.35 |
ENSMUST00000216289.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr2_+_172187485 | 1.33 |
ENSMUST00000028995.5
|
Fam210b
|
family with sequence similarity 210, member B |
| chr11_+_68979308 | 1.30 |
ENSMUST00000021273.13
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr11_-_82761954 | 1.29 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr5_-_137608886 | 1.29 |
ENSMUST00000142675.8
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr4_+_133097013 | 1.27 |
ENSMUST00000030669.8
|
Slc9a1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1 |
| chr8_-_86281946 | 1.26 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr16_-_4607751 | 1.24 |
ENSMUST00000117713.8
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr3_+_104545974 | 1.24 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr19_-_5416626 | 1.22 |
ENSMUST00000237167.2
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr2_-_30305779 | 1.21 |
ENSMUST00000102855.8
ENSMUST00000028207.13 |
Crat
|
carnitine acetyltransferase |
| chr5_+_24569802 | 1.21 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr18_-_43610829 | 1.21 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr3_+_96128427 | 1.20 |
ENSMUST00000090781.8
|
H2bc21
|
H2B clustered histone 21 |
| chr11_-_58213939 | 1.19 |
ENSMUST00000108829.8
|
Zfp672
|
zinc finger protein 672 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 31.3 | GO:0090089 | positive regulation of cellular pH reduction(GO:0032849) dipeptide transmembrane transport(GO:0035442) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) positive regulation of oligopeptide transport(GO:2000878) positive regulation of dipeptide transport(GO:2000880) regulation of dipeptide transmembrane transport(GO:2001148) positive regulation of dipeptide transmembrane transport(GO:2001150) |
| 6.5 | 19.6 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.3 | 27.3 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 2.2 | 38.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 2.2 | 35.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 1.6 | 6.4 | GO:0033366 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 1.6 | 4.8 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.5 | 6.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 1.5 | 14.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.4 | 13.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.4 | 18.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 1.3 | 13.9 | GO:0006113 | fermentation(GO:0006113) |
| 1.2 | 7.2 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 1.2 | 9.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 1.2 | 3.6 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 1.0 | 3.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 1.0 | 4.0 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.8 | 5.8 | GO:1903525 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.8 | 6.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.8 | 3.1 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.7 | 10.2 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.6 | 3.8 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.6 | 13.4 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.6 | 2.3 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.6 | 2.3 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.6 | 11.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.6 | 5.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.6 | 2.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.5 | 3.7 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.5 | 6.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.5 | 2.0 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.5 | 5.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.5 | 3.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 1.3 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.4 | 5.4 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.4 | 9.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 5.4 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 3.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 1.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 3.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 | 2.2 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.3 | 3.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.3 | 1.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 3.2 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.3 | 2.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.3 | 1.9 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 6.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 0.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 4.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 2.1 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.2 | 2.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 8.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.6 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 2.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 20.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 1.7 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 1.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 3.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 0.6 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.2 | 7.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 5.3 | GO:2000251 | actin crosslink formation(GO:0051764) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.0 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.2 | 3.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 2.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 3.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 3.0 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.2 | 5.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 4.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) histone H2A acetylation(GO:0043968) |
| 0.2 | 2.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.9 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 1.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 12.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.0 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 2.5 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 1.4 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 2.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 1.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 4.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.6 | GO:0034436 | glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 6.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 1.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.3 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.8 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.3 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.5 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.2 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.1 | 1.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 2.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.9 | GO:1901070 | guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.1 | 1.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 1.6 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 2.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 1.0 | GO:1901626 | regulation of postsynaptic membrane organization(GO:1901626) |
| 0.1 | 1.0 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.6 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 1.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 5.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 2.5 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 5.1 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 1.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 1.4 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 0.7 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.5 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 7.0 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.0 | 12.9 | GO:0051726 | regulation of cell cycle(GO:0051726) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 35.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.4 | 12.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.3 | 3.8 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.9 | 10.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.8 | 4.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.8 | 6.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.7 | 5.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.7 | 4.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.7 | 16.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 6.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 2.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.5 | 2.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.4 | 15.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 5.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.4 | 2.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 2.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 9.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 2.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 3.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 8.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 6.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 1.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 2.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 29.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 3.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 5.6 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.2 | 29.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.2 | 0.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 2.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 7.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 2.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 5.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 2.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 2.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 30.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 13.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 3.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 5.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 4.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 14.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 4.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.2 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.1 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.1 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 8.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 3.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 10.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 3.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 36.9 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 7.0 | 35.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 4.5 | 13.4 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 3.1 | 31.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 2.0 | 6.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 1.9 | 29.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.9 | 11.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.5 | 14.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.1 | 14.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 1.1 | 19.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 1.0 | 10.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.8 | 4.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.8 | 5.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.8 | 3.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.8 | 6.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.8 | 5.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 7.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.7 | 7.5 | GO:0070061 | fructose binding(GO:0070061) |
| 0.6 | 3.9 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.6 | 6.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.6 | 2.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.5 | 7.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 2.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.5 | 2.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.5 | 4.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.4 | 4.0 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.4 | 3.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.4 | 1.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.4 | 2.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.4 | 5.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.4 | 2.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.4 | 2.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.3 | 1.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 11.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 1.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 1.0 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 20.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 6.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 2.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 3.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 0.5 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.2 | 3.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 1.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 5.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 0.6 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 2.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 8.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 1.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 4.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.9 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 1.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 3.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 4.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 6.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 4.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 3.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 13.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 2.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 2.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 5.8 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 3.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 6.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 10.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 16.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 4.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 2.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 2.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 4.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.8 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 3.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 13.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.0 | 80.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 23.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 19.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 16.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 4.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 5.6 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 6.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 5.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 4.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 4.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 8.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 35.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.6 | 13.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.6 | 14.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.6 | 22.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 3.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 5.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 6.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 3.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 8.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.3 | 4.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 6.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 11.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 11.7 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.2 | 28.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 5.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 7.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 6.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.5 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 1.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 8.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 5.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 5.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |