Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
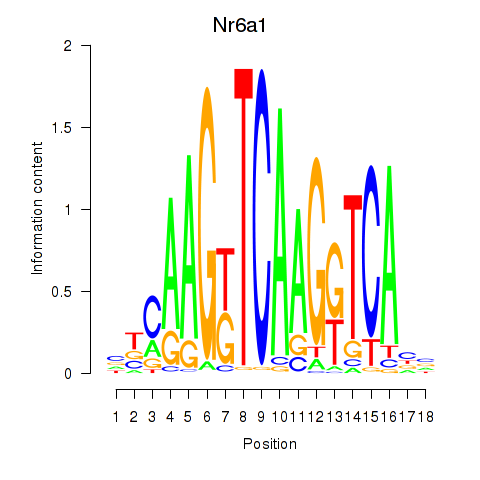
Results for Nr6a1
Z-value: 1.63
Transcription factors associated with Nr6a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr6a1
|
ENSMUSG00000063972.14 | Nr6a1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr6a1 | mm39_v1_chr2_-_38816229_38816366 | 0.21 | 2.2e-01 | Click! |
Activity profile of Nr6a1 motif
Sorted Z-values of Nr6a1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr6a1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_22769844 | 10.87 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_22769822 | 10.80 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr17_-_57535003 | 7.85 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr7_-_105249308 | 7.55 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr6_+_42222841 | 6.50 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr1_+_160806241 | 6.23 |
ENSMUST00000195760.2
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr1_+_160806194 | 6.20 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr3_+_107137924 | 5.84 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr9_-_118986123 | 5.49 |
ENSMUST00000010795.5
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr6_+_90310252 | 5.08 |
ENSMUST00000046128.12
ENSMUST00000164761.6 |
Uroc1
|
urocanase domain containing 1 |
| chr15_+_31572251 | 3.93 |
ENSMUST00000161088.3
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr2_+_24944367 | 3.89 |
ENSMUST00000100334.11
ENSMUST00000152122.8 ENSMUST00000116574.10 ENSMUST00000006646.15 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr2_+_24944407 | 3.82 |
ENSMUST00000102931.11
ENSMUST00000074422.14 ENSMUST00000132172.8 ENSMUST00000114388.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr14_+_79086665 | 3.80 |
ENSMUST00000227255.2
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr14_+_79086492 | 3.64 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr5_-_108822619 | 3.08 |
ENSMUST00000119270.2
ENSMUST00000163328.8 ENSMUST00000136227.2 |
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr11_-_60702081 | 3.06 |
ENSMUST00000018744.15
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr6_-_124613044 | 2.90 |
ENSMUST00000068797.3
ENSMUST00000218020.2 |
C1s2
|
complement component 1, s subcomponent 2 |
| chr12_-_28673259 | 2.72 |
ENSMUST00000220836.2
|
Colec11
|
collectin sub-family member 11 |
| chr12_-_28673311 | 2.64 |
ENSMUST00000036136.9
|
Colec11
|
collectin sub-family member 11 |
| chr1_+_131898325 | 2.31 |
ENSMUST00000027695.8
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr4_-_82423511 | 2.25 |
ENSMUST00000050872.15
ENSMUST00000064770.9 |
Nfib
|
nuclear factor I/B |
| chr7_+_44890497 | 2.24 |
ENSMUST00000213347.2
|
Slc6a16
|
solute carrier family 6, member 16 |
| chr4_+_138161958 | 2.17 |
ENSMUST00000044058.11
ENSMUST00000105813.8 ENSMUST00000105815.2 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr10_+_21854540 | 2.11 |
ENSMUST00000142174.8
ENSMUST00000164659.8 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr9_+_122634589 | 2.09 |
ENSMUST00000052740.14
|
Tcaim
|
T cell activation inhibitor, mitochondrial |
| chr17_-_26004507 | 1.86 |
ENSMUST00000140738.8
ENSMUST00000145053.2 ENSMUST00000138759.8 ENSMUST00000133071.8 ENSMUST00000077938.10 |
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr6_-_57512355 | 1.82 |
ENSMUST00000042766.6
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr5_-_36641456 | 1.70 |
ENSMUST00000119916.2
ENSMUST00000031097.8 |
Tada2b
|
transcriptional adaptor 2B |
| chr11_-_100661762 | 1.70 |
ENSMUST00000139341.2
ENSMUST00000017891.14 |
Ghdc
|
GH3 domain containing |
| chr9_-_50515089 | 1.69 |
ENSMUST00000000175.6
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr2_+_127267069 | 1.68 |
ENSMUST00000062211.4
|
Gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr2_+_32496957 | 1.68 |
ENSMUST00000113290.8
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr13_-_25204272 | 1.52 |
ENSMUST00000021772.4
|
Mrs2
|
MRS2 magnesium transporter |
| chr4_-_82423985 | 1.51 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr10_-_68377672 | 1.49 |
ENSMUST00000020103.9
|
Cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr13_+_31740117 | 1.47 |
ENSMUST00000042118.11
|
Foxq1
|
forkhead box Q1 |
| chr4_-_45530330 | 1.45 |
ENSMUST00000061986.12
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr2_+_32496990 | 1.44 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr5_+_8943943 | 1.39 |
ENSMUST00000196067.2
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr17_-_26004298 | 1.36 |
ENSMUST00000150324.8
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr8_-_110419867 | 1.30 |
ENSMUST00000034164.6
|
Ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr4_-_82423944 | 1.29 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr17_-_46956054 | 1.28 |
ENSMUST00000003642.7
|
Klc4
|
kinesin light chain 4 |
| chr18_-_6135888 | 1.19 |
ENSMUST00000182383.8
ENSMUST00000062584.14 ENSMUST00000077128.13 ENSMUST00000182038.2 ENSMUST00000182213.8 ENSMUST00000182559.8 |
Arhgap12
|
Rho GTPase activating protein 12 |
| chr5_-_137609691 | 1.13 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_-_137609634 | 0.94 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_-_137608886 | 0.94 |
ENSMUST00000142675.8
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr7_-_132178101 | 0.90 |
ENSMUST00000084500.8
|
Oat
|
ornithine aminotransferase |
| chr12_-_114752425 | 0.87 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr18_+_65020760 | 0.83 |
ENSMUST00000235343.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr11_-_76654375 | 0.82 |
ENSMUST00000010536.9
|
Gosr1
|
golgi SNAP receptor complex member 1 |
| chr17_+_73225292 | 0.75 |
ENSMUST00000024857.14
|
Lbh
|
limb-bud and heart |
| chr11_+_83742961 | 0.75 |
ENSMUST00000146786.8
|
Hnf1b
|
HNF1 homeobox B |
| chr18_+_65020910 | 0.74 |
ENSMUST00000236736.2
ENSMUST00000235743.2 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr12_-_115884332 | 0.73 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr18_+_65020885 | 0.70 |
ENSMUST00000236209.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr2_+_179713586 | 0.69 |
ENSMUST00000108901.8
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr12_+_44268134 | 0.68 |
ENSMUST00000122902.8
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr12_-_115916604 | 0.65 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr9_+_53212871 | 0.63 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr9_-_121668527 | 0.62 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr12_-_114547622 | 0.61 |
ENSMUST00000193893.6
ENSMUST00000103498.3 |
Ighv1-9
|
immunoglobulin heavy variable V1-9 |
| chrX_-_7884688 | 0.55 |
ENSMUST00000033503.3
|
Glod5
|
glyoxalase domain containing 5 |
| chr19_+_46695889 | 0.53 |
ENSMUST00000003655.9
|
As3mt
|
arsenite methyltransferase |
| chr5_+_36641922 | 0.51 |
ENSMUST00000060100.3
|
Ccdc96
|
coiled-coil domain containing 96 |
| chr5_+_137015873 | 0.41 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr14_+_76725876 | 0.41 |
ENSMUST00000101618.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr12_-_114672701 | 0.39 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr6_+_21985902 | 0.38 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr9_+_45311000 | 0.37 |
ENSMUST00000216289.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr5_+_92524813 | 0.35 |
ENSMUST00000120781.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr12_-_114646685 | 0.33 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr5_+_30437579 | 0.33 |
ENSMUST00000145167.9
|
Selenoi
|
selenoprotein I |
| chr12_-_113896002 | 0.32 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr5_-_137015683 | 0.30 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr12_-_114878652 | 0.26 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr18_+_65158873 | 0.24 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr16_+_10297923 | 0.21 |
ENSMUST00000230395.2
|
Ciita
|
class II transactivator |
| chr5_+_129802127 | 0.21 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr13_+_113931038 | 0.20 |
ENSMUST00000091201.7
|
Arl15
|
ADP-ribosylation factor-like 15 |
| chr16_+_11140779 | 0.18 |
ENSMUST00000115814.4
|
Snx29
|
sorting nexin 29 |
| chr10_+_23846604 | 0.16 |
ENSMUST00000092659.4
|
Taar5
|
trace amine-associated receptor 5 |
| chr1_+_151304293 | 0.16 |
ENSMUST00000065625.12
|
Trmt1l
|
tRNA methyltransferase 1 like |
| chr7_+_16678568 | 0.16 |
ENSMUST00000094807.6
|
Pnmal2
|
PNMA-like 2 |
| chr19_+_47843461 | 0.15 |
ENSMUST00000237121.2
|
Gsto1
|
glutathione S-transferase omega 1 |
| chr13_+_25239947 | 0.13 |
ENSMUST00000036932.15
|
Dcdc2a
|
doublecortin domain containing 2a |
| chr15_-_98063441 | 0.12 |
ENSMUST00000123922.2
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr2_+_157266175 | 0.09 |
ENSMUST00000029175.14
ENSMUST00000092576.11 |
Src
|
Rous sarcoma oncogene |
| chr1_+_171156942 | 0.08 |
ENSMUST00000111299.8
ENSMUST00000064950.11 |
Dedd
|
death effector domain-containing |
| chr14_+_58310143 | 0.08 |
ENSMUST00000022545.14
|
Fgf9
|
fibroblast growth factor 9 |
| chr12_-_71183371 | 0.07 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr1_+_171156568 | 0.05 |
ENSMUST00000111300.8
|
Dedd
|
death effector domain-containing |
| chr15_+_99936516 | 0.04 |
ENSMUST00000100203.10
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr5_-_123711098 | 0.03 |
ENSMUST00000031388.13
|
Vps33a
|
VPS33A CORVET/HOPS core subunit |
| chr12_+_78240999 | 0.02 |
ENSMUST00000211288.3
|
Gm6657
|
predicted gene 6657 |
| chr12_-_114579763 | 0.02 |
ENSMUST00000103500.2
|
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.7 | GO:0097037 | heme export(GO:0097037) |
| 2.0 | 7.9 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.9 | 7.6 | GO:0015886 | heme transport(GO:0015886) |
| 1.6 | 12.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.3 | 7.7 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 1.3 | 5.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.0 | 3.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 1.0 | 5.1 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.6 | 2.3 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 | 1.4 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.4 | 2.2 | GO:1904925 | negative regulation of mitochondrial fusion(GO:0010637) negative regulation of defense response to virus by host(GO:0050689) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.3 | 3.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 1.3 | GO:0009838 | abscission(GO:0009838) |
| 0.3 | 3.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 3.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.7 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.2 | 2.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 2.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 0.7 | GO:0035565 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.2 | 1.7 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 1.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 2.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.9 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.8 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.1 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 6.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.2 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.8 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 1.7 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.7 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.5 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.2 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 3.0 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.2 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.7 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.4 | 1.7 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 5.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 7.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 27.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 5.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 8.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.0 | 3.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.6 | 2.3 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.6 | 1.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.4 | 3.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 5.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 6.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 2.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 1.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.2 | 33.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 2.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 5.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 2.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 5.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 7.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 7.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 3.0 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0045174 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 6.5 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 1.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 12.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 21.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 12.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.6 | 7.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 5.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 21.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 3.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 6.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |