Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
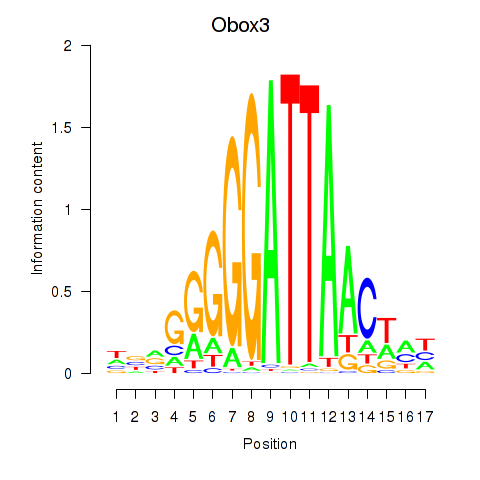
Results for Obox3
Z-value: 1.01
Transcription factors associated with Obox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Obox3
|
ENSMUSG00000066772.13 | Obox3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Obox3 | mm39_v1_chr7_-_15361801_15361801 | 0.37 | 2.6e-02 | Click! |
Activity profile of Obox3 motif
Sorted Z-values of Obox3 motif
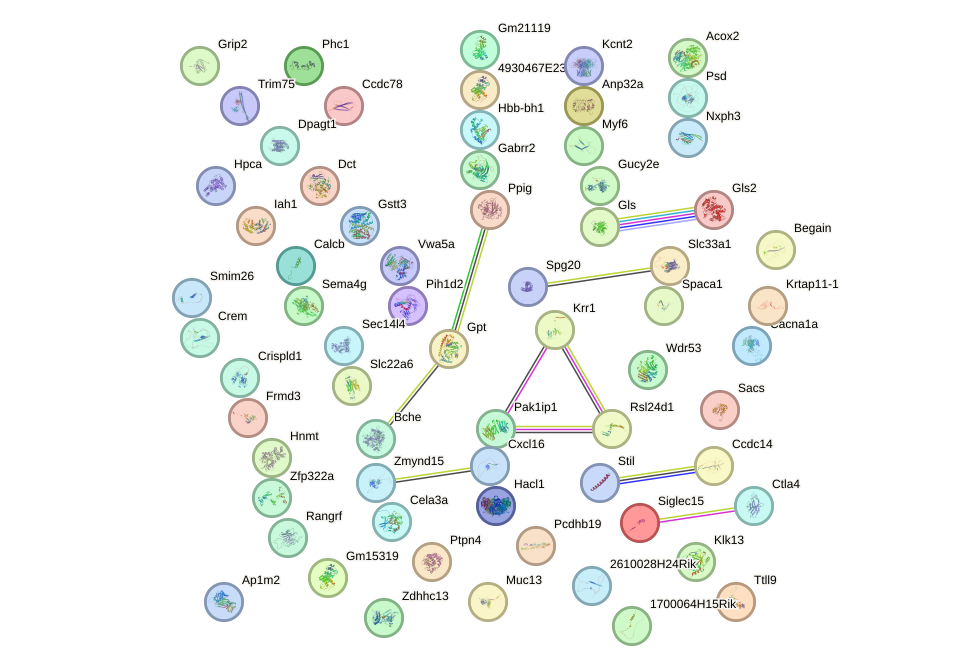
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_76579885 | 3.27 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr15_+_76579960 | 3.08 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr10_-_75617245 | 2.45 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr11_+_3981769 | 1.45 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr2_-_23939401 | 1.36 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr3_-_63872189 | 0.84 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr4_-_137137088 | 0.80 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr7_-_103492361 | 0.72 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr3_-_63872079 | 0.71 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr11_-_68864666 | 0.70 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr10_+_111808569 | 0.65 |
ENSMUST00000163048.8
ENSMUST00000174653.2 |
Krr1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr2_+_144435974 | 0.62 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr6_-_122317156 | 0.58 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chrX_+_73468140 | 0.57 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr9_+_62249341 | 0.55 |
ENSMUST00000135395.8
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr7_+_43361930 | 0.49 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr3_-_73615732 | 0.45 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr9_+_62249730 | 0.44 |
ENSMUST00000156461.2
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chrX_+_141010919 | 0.39 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr1_+_140173787 | 0.37 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr2_-_164455520 | 0.35 |
ENSMUST00000094351.11
ENSMUST00000109338.2 |
Wfdc8
|
WAP four-disulfide core domain 8 |
| chr2_+_152804405 | 0.35 |
ENSMUST00000099197.9
ENSMUST00000103155.10 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr2_+_69553141 | 0.34 |
ENSMUST00000090858.10
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chrX_+_139565319 | 0.30 |
ENSMUST00000128809.2
|
Mid2
|
midline 2 |
| chr6_-_91784299 | 0.30 |
ENSMUST00000159684.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr19_-_46314945 | 0.30 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr18_-_78100610 | 0.29 |
ENSMUST00000170760.3
|
Siglec15
|
sialic acid binding Ig-like lectin 15 |
| chr18_+_42644552 | 0.28 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr3_-_19682833 | 0.28 |
ENSMUST00000119133.2
|
1700064H15Rik
|
RIKEN cDNA 1700064H15 gene |
| chr6_-_91784405 | 0.28 |
ENSMUST00000162300.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr17_+_9207165 | 0.27 |
ENSMUST00000024650.12
|
1700010I14Rik
|
RIKEN cDNA 1700010I14 gene |
| chr11_+_70350963 | 0.27 |
ENSMUST00000126105.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr10_+_76284907 | 0.26 |
ENSMUST00000092406.12
|
2610028H24Rik
|
RIKEN cDNA 2610028H24 gene |
| chr11_+_70350725 | 0.26 |
ENSMUST00000147289.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr1_+_17797257 | 0.25 |
ENSMUST00000159958.8
ENSMUST00000160305.8 ENSMUST00000095075.5 |
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr7_+_48438751 | 0.25 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr16_-_20135500 | 0.24 |
ENSMUST00000182741.2
|
Cyp2ab1
|
cytochrome P450, family 2, subfamily ab, polypeptide 1 |
| chr7_+_114318746 | 0.24 |
ENSMUST00000182044.2
|
Calcb
|
calcitonin-related polypeptide, beta |
| chr4_-_129015493 | 0.23 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
| chr11_+_60956624 | 0.23 |
ENSMUST00000041944.9
ENSMUST00000108717.3 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr19_+_3817396 | 0.23 |
ENSMUST00000052699.13
ENSMUST00000113974.11 ENSMUST00000113972.9 ENSMUST00000113973.8 ENSMUST00000113977.9 ENSMUST00000113968.9 |
Kmt5b
|
lysine methyltransferase 5B |
| chr14_-_118289557 | 0.22 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr12_-_109019507 | 0.22 |
ENSMUST00000185745.2
ENSMUST00000239108.2 |
Begain
|
brain-enriched guanylate kinase-associated |
| chr4_+_73931679 | 0.22 |
ENSMUST00000098006.9
ENSMUST00000084474.6 |
Frmd3
|
FERM domain containing 3 |
| chr11_-_95405368 | 0.21 |
ENSMUST00000058866.8
|
Nxph3
|
neurexophilin 3 |
| chr14_+_61375893 | 0.21 |
ENSMUST00000089394.10
|
Sacs
|
sacsin |
| chr17_+_9207180 | 0.20 |
ENSMUST00000151609.2
ENSMUST00000232775.2 ENSMUST00000136954.3 |
1700010I14Rik
|
RIKEN cDNA 1700010I14 gene |
| chr9_+_73020708 | 0.20 |
ENSMUST00000169399.8
ENSMUST00000034738.14 |
Rsl24d1
|
ribosomal L24 domain containing 1 |
| chr17_+_26005538 | 0.20 |
ENSMUST00000095500.5
|
Ccdc78
|
coiled-coil domain containing 78 |
| chr11_-_69127848 | 0.18 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr8_-_65440483 | 0.18 |
ENSMUST00000210982.2
|
Trim75
|
tripartite motif-containing 75 |
| chr13_-_23553327 | 0.18 |
ENSMUST00000125328.2
ENSMUST00000145451.8 ENSMUST00000050101.9 |
Zfp322a
|
zinc finger protein 322A |
| chr11_+_51895166 | 0.17 |
ENSMUST00000109076.2
|
Cdkl3
|
cyclin-dependent kinase-like 3 |
| chr1_-_52271455 | 0.17 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr1_+_60948149 | 0.17 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr10_-_107330580 | 0.17 |
ENSMUST00000044210.5
|
Myf6
|
myogenic factor 6 |
| chr5_-_93688059 | 0.16 |
ENSMUST00000200745.2
|
Pramel33
|
PRAME like 33 |
| chr16_+_34511073 | 0.16 |
ENSMUST00000231609.2
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr4_-_34050076 | 0.16 |
ENSMUST00000029927.6
|
Spaca1
|
sperm acrosome associated 1 |
| chr19_+_3817953 | 0.16 |
ENSMUST00000113970.8
|
Kmt5b
|
lysine methyltransferase 5B |
| chr4_-_34050038 | 0.16 |
ENSMUST00000084734.11
|
Spaca1
|
sperm acrosome associated 1 |
| chr1_+_60948307 | 0.15 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr18_+_37630044 | 0.15 |
ENSMUST00000059571.7
|
Pcdhb19
|
protocadherin beta 19 |
| chr8_+_20078070 | 0.15 |
ENSMUST00000084046.6
|
Gm15319
|
predicted gene 15319 |
| chr8_+_21154935 | 0.15 |
ENSMUST00000178438.3
|
Gm21119
|
predicted gene, 21119 |
| chr8_+_19779592 | 0.15 |
ENSMUST00000098909.5
|
4930467E23Rik
|
RIKEN cDNA 4930467E23 gene |
| chr3_-_73615535 | 0.14 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
| chr14_+_14210932 | 0.14 |
ENSMUST00000022271.14
|
Acox2
|
acyl-Coenzyme A oxidase 2, branched chain |
| chr16_+_32066037 | 0.14 |
ENSMUST00000141820.8
ENSMUST00000178573.8 ENSMUST00000023474.4 ENSMUST00000135289.2 |
Wdr53
|
WD repeat domain 53 |
| chr8_-_65440298 | 0.13 |
ENSMUST00000095295.3
|
Trim75
|
tripartite motif-containing 75 |
| chr13_+_41154478 | 0.13 |
ENSMUST00000046951.10
|
Pak1ip1
|
PAK1 interacting protein 1 |
| chr19_+_3818112 | 0.12 |
ENSMUST00000005518.16
ENSMUST00000237440.2 ENSMUST00000152935.8 ENSMUST00000176262.8 ENSMUST00000176407.8 ENSMUST00000176926.8 ENSMUST00000176512.8 |
Kmt5b
|
lysine methyltransferase 5B |
| chr7_-_11414074 | 0.12 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr14_-_31362835 | 0.11 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr6_+_90246088 | 0.11 |
ENSMUST00000058039.3
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr16_-_17732923 | 0.10 |
ENSMUST00000012279.6
ENSMUST00000232493.2 |
Gsc2
|
goosecoid homebox 2 |
| chr12_-_101943134 | 0.10 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr1_-_119765343 | 0.09 |
ENSMUST00000064091.12
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr3_+_55023594 | 0.09 |
ENSMUST00000146109.2
|
Spg20
|
spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) |
| chr4_+_33062999 | 0.09 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr9_-_40025502 | 0.08 |
ENSMUST00000217536.3
|
Olfr984
|
olfactory receptor 984 |
| chr12_+_21366386 | 0.08 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chr4_+_114857370 | 0.08 |
ENSMUST00000129957.8
|
Stil
|
Scl/Tal1 interrupting locus |
| chr1_-_119765068 | 0.07 |
ENSMUST00000163435.8
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr9_+_38630317 | 0.06 |
ENSMUST00000129598.2
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr1_-_119764729 | 0.05 |
ENSMUST00000163621.2
ENSMUST00000168303.8 |
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr18_-_3281752 | 0.05 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr9_-_21223631 | 0.03 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr18_-_43526411 | 0.03 |
ENSMUST00000025379.14
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr19_+_8595369 | 0.03 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr7_-_110581376 | 0.02 |
ENSMUST00000154466.2
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr16_-_19341016 | 0.02 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr15_+_81548090 | 0.02 |
ENSMUST00000023029.15
ENSMUST00000174229.8 ENSMUST00000172748.8 |
L3mbtl2
|
L3MBTL2 polycomb repressive complex 1 subunit |
| chr16_+_33614378 | 0.02 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr9_+_50528608 | 0.00 |
ENSMUST00000000171.15
ENSMUST00000151197.8 |
Pih1d2
|
PIH1 domain containing 2 |
| chr19_+_44980565 | 0.00 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.6 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 0.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 0.6 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 2.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 1.5 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 1.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.3 | 1.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 0.6 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 0.7 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 2.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |