Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Olig2_Olig3
Z-value: 1.17
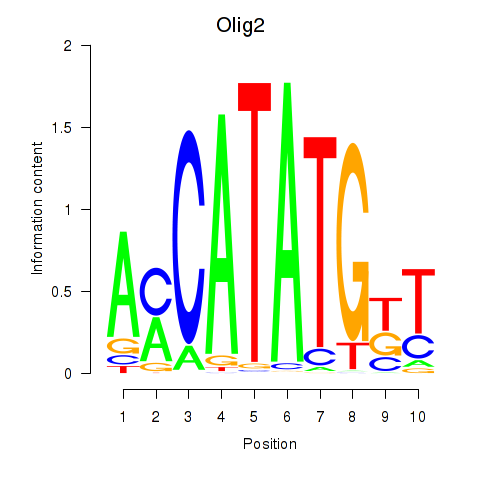
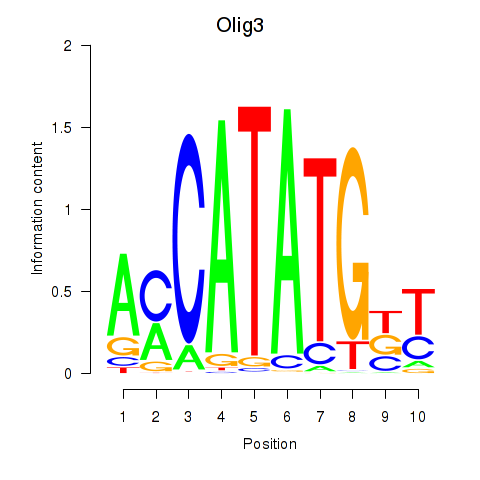
Transcription factors associated with Olig2_Olig3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Olig2
|
ENSMUSG00000039830.10 | Olig2 |
|
Olig3
|
ENSMUSG00000045591.7 | Olig3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Olig3 | mm39_v1_chr10_+_19232281_19232312 | -0.28 | 1.0e-01 | Click! |
| Olig2 | mm39_v1_chr16_+_91022300_91022345 | 0.07 | 6.6e-01 | Click! |
Activity profile of Olig2_Olig3 motif
Sorted Z-values of Olig2_Olig3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Olig2_Olig3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_121983720 | 8.78 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr19_+_20579322 | 7.39 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr10_+_87697155 | 6.03 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr6_+_121815473 | 5.82 |
ENSMUST00000032228.9
|
Mug1
|
murinoglobulin 1 |
| chr6_-_41291634 | 5.74 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr6_+_124489364 | 5.43 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr6_+_41331039 | 5.35 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr10_-_127843377 | 5.12 |
ENSMUST00000219447.2
ENSMUST00000219780.2 ENSMUST00000219707.2 ENSMUST00000219953.2 ENSMUST00000219183.2 |
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr5_-_147259245 | 4.90 |
ENSMUST00000100433.5
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr9_-_121745354 | 4.55 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr5_-_87485023 | 4.15 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr3_+_20039775 | 4.13 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr1_+_160806241 | 3.89 |
ENSMUST00000195760.2
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr9_-_48516447 | 3.58 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr15_-_75963446 | 3.12 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr11_-_69696428 | 3.03 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr17_-_32639936 | 3.01 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr18_+_12776358 | 2.79 |
ENSMUST00000234966.2
ENSMUST00000025294.9 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr8_+_86219191 | 2.74 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr4_-_137157824 | 2.66 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr8_-_106660470 | 2.62 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr7_+_130633776 | 2.57 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr9_-_72892617 | 2.52 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr16_+_22738987 | 2.44 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr6_+_90439596 | 2.38 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr6_+_90439544 | 2.35 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr1_-_180027151 | 2.35 |
ENSMUST00000161743.3
|
Coq8a
|
coenzyme Q8A |
| chr15_-_76010736 | 2.30 |
ENSMUST00000054022.12
ENSMUST00000089654.4 |
BC024139
|
cDNA sequence BC024139 |
| chr9_-_110571645 | 2.27 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr3_-_86906591 | 2.26 |
ENSMUST00000063869.11
ENSMUST00000029717.4 |
Cd1d1
|
CD1d1 antigen |
| chr2_-_25391729 | 2.22 |
ENSMUST00000015227.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr17_-_31363245 | 2.22 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr2_+_155224105 | 2.22 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr11_+_48729499 | 2.18 |
ENSMUST00000129674.2
|
Trim7
|
tripartite motif-containing 7 |
| chr7_+_37882642 | 2.13 |
ENSMUST00000178207.10
ENSMUST00000179525.10 |
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr8_+_95564949 | 2.12 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr5_-_104125192 | 1.97 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr3_-_107851021 | 1.94 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr5_-_104125226 | 1.89 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_104125270 | 1.86 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr14_+_30376310 | 1.85 |
ENSMUST00000064230.16
|
Rft1
|
RFT1 homolog |
| chr3_-_113367891 | 1.85 |
ENSMUST00000142505.9
|
Amy1
|
amylase 1, salivary |
| chr1_+_88093726 | 1.80 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr14_-_66191177 | 1.79 |
ENSMUST00000042046.5
|
Scara3
|
scavenger receptor class A, member 3 |
| chr11_-_73215442 | 1.78 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr1_-_139786421 | 1.77 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr19_-_20704896 | 1.75 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr10_-_95678786 | 1.74 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr5_+_90608751 | 1.71 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr12_+_87194476 | 1.69 |
ENSMUST00000063117.10
ENSMUST00000220574.2 |
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr1_+_160806194 | 1.67 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr1_-_185849448 | 1.67 |
ENSMUST00000045388.8
|
Lyplal1
|
lysophospholipase-like 1 |
| chr17_-_26011357 | 1.60 |
ENSMUST00000236683.2
|
Antkmt
|
adenine nucleotide translocase lysine methyltransferase |
| chr1_-_139487951 | 1.59 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chrM_+_5319 | 1.58 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr10_-_95678748 | 1.56 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr2_-_52448552 | 1.56 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr8_-_85620537 | 1.44 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr15_+_100202079 | 1.43 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr3_+_146302832 | 1.43 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr18_+_36797113 | 1.41 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr15_+_100202021 | 1.36 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr10_+_110756031 | 1.35 |
ENSMUST00000220409.2
ENSMUST00000219502.2 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr9_+_72892850 | 1.34 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr3_-_37473715 | 1.26 |
ENSMUST00000138949.2
ENSMUST00000149449.8 ENSMUST00000108118.9 ENSMUST00000108117.3 ENSMUST00000099130.9 ENSMUST00000052645.13 |
Nudt6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr9_+_89093210 | 1.23 |
ENSMUST00000118870.8
ENSMUST00000085256.8 |
Mthfs
|
5, 10-methenyltetrahydrofolate synthetase |
| chr2_+_136733421 | 1.19 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr7_-_44465998 | 1.18 |
ENSMUST00000209072.2
ENSMUST00000047356.11 |
Atf5
|
activating transcription factor 5 |
| chr4_-_137137088 | 1.17 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr11_+_115494751 | 1.15 |
ENSMUST00000058109.9
|
Mrps7
|
mitchondrial ribosomal protein S7 |
| chr17_+_47083561 | 1.15 |
ENSMUST00000071430.7
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chr14_+_74969737 | 1.12 |
ENSMUST00000022573.17
ENSMUST00000175712.8 ENSMUST00000176957.8 |
Esd
|
esterase D/formylglutathione hydrolase |
| chr14_+_40873399 | 1.09 |
ENSMUST00000225792.2
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr10_+_3490232 | 1.07 |
ENSMUST00000019896.5
|
Iyd
|
iodotyrosine deiodinase |
| chr17_+_35345292 | 1.07 |
ENSMUST00000061859.7
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr6_+_113449237 | 1.07 |
ENSMUST00000204447.3
|
Il17rc
|
interleukin 17 receptor C |
| chr7_+_127400016 | 1.06 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_-_136733282 | 1.04 |
ENSMUST00000028730.13
ENSMUST00000110089.9 ENSMUST00000227806.2 |
Mkks
ENSMUSG00000115423.3
|
McKusick-Kaufman syndrome novel protein |
| chr16_-_45830575 | 1.03 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr4_+_106924181 | 1.01 |
ENSMUST00000106758.8
ENSMUST00000145324.8 ENSMUST00000106760.8 |
Cyb5rl
|
cytochrome b5 reductase-like |
| chr12_-_87194658 | 1.00 |
ENSMUST00000037788.6
|
Pomt2
|
protein-O-mannosyltransferase 2 |
| chr17_+_34372046 | 0.99 |
ENSMUST00000114232.4
|
H2-DMb1
|
histocompatibility 2, class II, locus Mb1 |
| chr1_-_74343543 | 0.99 |
ENSMUST00000016309.16
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr3_+_122688721 | 0.98 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr7_-_99340830 | 0.97 |
ENSMUST00000208713.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr9_-_88601866 | 0.95 |
ENSMUST00000113110.5
|
Mthfsl
|
5, 10-methenyltetrahydrofolate synthetase-like |
| chr2_+_59442378 | 0.94 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr4_+_141473983 | 0.94 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr7_+_127399848 | 0.94 |
ENSMUST00000139068.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr15_+_7159038 | 0.94 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr1_-_80439165 | 0.93 |
ENSMUST00000211023.2
|
Gm45261
|
predicted gene 45261 |
| chr6_-_122317156 | 0.93 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr1_+_167445815 | 0.91 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr4_+_133246274 | 0.90 |
ENSMUST00000149807.2
ENSMUST00000042919.16 ENSMUST00000153811.2 ENSMUST00000105901.2 ENSMUST00000121797.2 |
Kdf1
|
keratinocyte differentiation factor 1 |
| chrX_+_140258381 | 0.90 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr11_-_53321242 | 0.87 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr18_-_46444804 | 0.87 |
ENSMUST00000236934.2
|
Ccdc112
|
coiled-coil domain containing 112 |
| chr8_+_36956345 | 0.86 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr11_+_32592707 | 0.83 |
ENSMUST00000109366.8
ENSMUST00000093205.13 ENSMUST00000076383.8 |
Fbxw11
|
F-box and WD-40 domain protein 11 |
| chr7_-_126062272 | 0.80 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr9_-_59260713 | 0.79 |
ENSMUST00000026265.8
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr8_-_34614187 | 0.78 |
ENSMUST00000033910.9
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr18_-_35760260 | 0.77 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr3_-_57202301 | 0.76 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr15_-_60793115 | 0.76 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr7_-_44711771 | 0.76 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr9_-_26910833 | 0.75 |
ENSMUST00000060513.8
ENSMUST00000120367.8 |
Acad8
|
acyl-Coenzyme A dehydrogenase family, member 8 |
| chr7_+_27290969 | 0.75 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr4_+_85972125 | 0.75 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr16_+_17149235 | 0.75 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr9_+_37313193 | 0.74 |
ENSMUST00000214185.3
|
Robo4
|
roundabout guidance receptor 4 |
| chr7_-_139162706 | 0.73 |
ENSMUST00000106095.3
|
Nkx6-2
|
NK6 homeobox 2 |
| chr10_-_127358231 | 0.73 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr10_-_127358300 | 0.72 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr11_+_43046476 | 0.72 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr10_+_3316057 | 0.72 |
ENSMUST00000043374.7
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr9_+_37313287 | 0.72 |
ENSMUST00000115048.10
ENSMUST00000115046.9 ENSMUST00000102895.7 ENSMUST00000239486.2 |
Robo4
|
roundabout guidance receptor 4 |
| chr5_-_25107313 | 0.72 |
ENSMUST00000131486.2
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr4_+_106924209 | 0.71 |
ENSMUST00000154283.2
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr7_-_45546081 | 0.70 |
ENSMUST00000120299.4
ENSMUST00000039049.16 |
Syngr4
|
synaptogyrin 4 |
| chr7_+_44499374 | 0.66 |
ENSMUST00000141311.8
ENSMUST00000107880.9 ENSMUST00000208384.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr8_-_107792264 | 0.65 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr6_+_72332423 | 0.65 |
ENSMUST00000069695.9
ENSMUST00000132243.3 |
Tmem150a
|
transmembrane protein 150A |
| chr11_+_102036356 | 0.64 |
ENSMUST00000055409.6
|
Nags
|
N-acetylglutamate synthase |
| chr4_-_150087587 | 0.63 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr8_-_107064615 | 0.63 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr5_-_24652775 | 0.62 |
ENSMUST00000123167.2
ENSMUST00000030799.15 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr2_+_65499097 | 0.61 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chrY_+_2900989 | 0.61 |
ENSMUST00000187842.7
|
Gm10352
|
predicted gene 10352 |
| chrY_+_3771673 | 0.61 |
ENSMUST00000186140.7
|
Gm3376
|
predicted gene 3376 |
| chrY_-_3306449 | 0.61 |
ENSMUST00000189592.7
|
Gm21677
|
predicted gene, 21677 |
| chrY_-_3378783 | 0.61 |
ENSMUST00000187277.7
|
Gm21704
|
predicted gene, 21704 |
| chrY_-_2796205 | 0.61 |
ENSMUST00000187482.2
|
Gm4064
|
predicted gene 4064 |
| chrY_+_2862139 | 0.61 |
ENSMUST00000189964.7
ENSMUST00000188114.2 |
Gm10256
|
predicted gene 10256 |
| chrY_+_2830680 | 0.61 |
ENSMUST00000171534.8
ENSMUST00000100360.5 ENSMUST00000179404.8 |
Rbmy
Gm10256
|
RNA binding motif protein, Y chromosome predicted gene 10256 |
| chr17_+_56935118 | 0.60 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chr10_+_78410180 | 0.59 |
ENSMUST00000218061.2
ENSMUST00000218787.2 ENSMUST00000105384.5 ENSMUST00000218875.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr10_-_117582259 | 0.59 |
ENSMUST00000079041.7
|
Slc35e3
|
solute carrier family 35, member E3 |
| chr2_+_160730019 | 0.58 |
ENSMUST00000109455.9
ENSMUST00000040872.13 |
Lpin3
|
lipin 3 |
| chr15_+_30172716 | 0.58 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr6_+_121709891 | 0.57 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr2_-_65397809 | 0.57 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr3_+_105821450 | 0.57 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr1_+_153300874 | 0.57 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr7_-_103320398 | 0.56 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr1_+_13738967 | 0.55 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr2_+_25293140 | 0.55 |
ENSMUST00000154809.8
ENSMUST00000055921.14 ENSMUST00000141567.8 |
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr11_-_109986804 | 0.55 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr16_-_44153288 | 0.55 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr2_+_118603247 | 0.54 |
ENSMUST00000061360.4
ENSMUST00000130293.8 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chrY_-_3410148 | 0.53 |
ENSMUST00000190283.7
ENSMUST00000188091.7 ENSMUST00000169382.3 |
Gm21708
Gm21704
|
predicted gene, 21708 predicted gene, 21704 |
| chr5_-_5529119 | 0.53 |
ENSMUST00000115447.2
|
Pttg1ip2
|
PTTG1IP family member 2 |
| chr17_-_24439712 | 0.53 |
ENSMUST00000024930.8
|
Tedc2
|
tubulin epsilon and delta complex 2 |
| chr10_-_78131228 | 0.53 |
ENSMUST00000105387.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr2_+_65760477 | 0.52 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr11_-_69811717 | 0.51 |
ENSMUST00000152589.2
ENSMUST00000108612.8 ENSMUST00000108611.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr19_+_43770619 | 0.51 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr10_+_59715378 | 0.51 |
ENSMUST00000147914.8
ENSMUST00000146590.8 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr7_+_123061497 | 0.51 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr7_+_45546365 | 0.51 |
ENSMUST00000069772.16
ENSMUST00000210503.2 ENSMUST00000209350.2 |
Tmem143
|
transmembrane protein 143 |
| chr10_-_126877382 | 0.50 |
ENSMUST00000116231.4
|
Eef1akmt3
|
EEF1A lysine methyltransferase 3 |
| chr2_+_160730076 | 0.50 |
ENSMUST00000109457.3
|
Lpin3
|
lipin 3 |
| chr2_+_86655007 | 0.50 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr7_+_15853792 | 0.49 |
ENSMUST00000006178.5
|
Kptn
|
kaptin |
| chr10_-_79819821 | 0.49 |
ENSMUST00000124536.2
|
Tmem259
|
transmembrane protein 259 |
| chr3_-_127574167 | 0.49 |
ENSMUST00000029662.12
|
Alpk1
|
alpha-kinase 1 |
| chr7_+_44499005 | 0.49 |
ENSMUST00000150335.2
ENSMUST00000107882.8 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr13_+_92491234 | 0.49 |
ENSMUST00000022218.6
|
Dhfr
|
dihydrofolate reductase |
| chr3_-_146302343 | 0.48 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr6_+_97906760 | 0.48 |
ENSMUST00000101123.10
|
Mitf
|
melanogenesis associated transcription factor |
| chr2_-_132089667 | 0.48 |
ENSMUST00000110163.8
ENSMUST00000180286.2 ENSMUST00000028816.9 |
Tmem230
|
transmembrane protein 230 |
| chr6_+_48372771 | 0.47 |
ENSMUST00000114572.9
|
Krba1
|
KRAB-A domain containing 1 |
| chr4_+_11579648 | 0.47 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr2_-_34951443 | 0.46 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr14_+_50595361 | 0.46 |
ENSMUST00000185091.2
|
Tlr11
|
toll-like receptor 11 |
| chr5_-_66775979 | 0.46 |
ENSMUST00000162382.8
ENSMUST00000159786.8 ENSMUST00000162349.8 ENSMUST00000087256.12 ENSMUST00000160103.8 ENSMUST00000160870.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr1_+_135746330 | 0.45 |
ENSMUST00000038760.10
|
Lad1
|
ladinin |
| chr11_+_67665434 | 0.44 |
ENSMUST00000181566.2
|
Gsg1l2
|
GSG1-like 2 |
| chr15_+_75865604 | 0.44 |
ENSMUST00000089669.6
|
Mapk15
|
mitogen-activated protein kinase 15 |
| chr8_+_53964721 | 0.44 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr7_+_44498640 | 0.44 |
ENSMUST00000054343.15
ENSMUST00000142880.3 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr2_-_101479846 | 0.43 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chr9_+_24194729 | 0.43 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr2_+_155078522 | 0.43 |
ENSMUST00000150602.2
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr18_+_61044830 | 0.43 |
ENSMUST00000040359.6
|
Arsi
|
arylsulfatase i |
| chr5_+_124621521 | 0.43 |
ENSMUST00000111453.2
|
Snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr3_+_154302311 | 0.42 |
ENSMUST00000192462.6
ENSMUST00000029850.15 |
Cryz
|
crystallin, zeta |
| chr17_-_27386763 | 0.42 |
ENSMUST00000025046.4
|
Ip6k3
|
inositol hexaphosphate kinase 3 |
| chr10_+_3316505 | 0.42 |
ENSMUST00000217573.2
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr9_+_66853343 | 0.42 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr14_+_28233301 | 0.42 |
ENSMUST00000112272.2
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr2_+_155078449 | 0.41 |
ENSMUST00000109682.9
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr17_+_37504783 | 0.41 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr3_-_85653573 | 0.41 |
ENSMUST00000118408.8
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr6_+_5725639 | 0.41 |
ENSMUST00000115556.8
ENSMUST00000115555.8 ENSMUST00000115559.10 |
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr3_-_57202546 | 0.41 |
ENSMUST00000196506.2
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr7_-_103420801 | 0.40 |
ENSMUST00000106878.3
|
Olfr69
|
olfactory receptor 69 |
| chr13_+_67979225 | 0.40 |
ENSMUST00000221266.2
|
Gm10037
|
predicted gene 10037 |
| chr9_-_119654522 | 0.40 |
ENSMUST00000070617.8
|
Scn11a
|
sodium channel, voltage-gated, type XI, alpha |
| chr5_-_135991117 | 0.40 |
ENSMUST00000111150.2
ENSMUST00000054895.4 |
Ssc4d
|
scavenger receptor cysteine rich family, 4 domains |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.0 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.8 | 2.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.8 | 7.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.8 | 2.3 | GO:0048003 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.7 | 5.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.7 | 2.7 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.5 | 3.0 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.5 | 1.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 1.4 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.5 | 1.8 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.4 | 4.9 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.4 | 1.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.4 | 6.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 1.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.3 | 2.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 0.9 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 1.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 1.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 1.8 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 0.5 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.2 | 2.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 3.4 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 1.7 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.2 | GO:2000405 | negative regulation of T cell migration(GO:2000405) |
| 0.2 | 1.0 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.2 | 0.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 1.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 4.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 0.7 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.2 | 2.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 2.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.2 | 0.9 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 0.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 4.7 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 1.0 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 1.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 0.9 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.8 | GO:0071484 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.9 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.7 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.6 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 2.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 4.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.8 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.4 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 1.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.6 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.7 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 0.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.4 | GO:0000239 | pachytene(GO:0000239) meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.1 | 0.4 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 1.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.3 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 1.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.7 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.1 | 10.0 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 5.3 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 2.1 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.2 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 1.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.1 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.3 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.3 | GO:0034334 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) |
| 0.1 | 1.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 0.3 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.2 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 2.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.4 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.3 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.8 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 4.7 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.4 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 9.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.5 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.9 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 3.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 3.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.8 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 1.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0002606 | regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 3.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 2.2 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.3 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.7 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.6 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 1.8 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 0.4 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 1.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 2.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 0.9 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.2 | 2.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.4 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.7 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 21.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 7.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 3.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 2.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 4.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.7 | 2.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.6 | 5.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.6 | 1.8 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.6 | 1.7 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.5 | 2.7 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.5 | 3.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.5 | 2.0 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.5 | 1.4 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.4 | 4.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 2.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 2.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.3 | 1.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.6 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.2 | 0.9 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 0.8 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 6.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 21.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 6.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 2.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0004320 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 1.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.6 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 2.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 18.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.1 | 4.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.2 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.4 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 2.6 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.8 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.4 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.1 | 1.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 4.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.4 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 1.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 1.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 2.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 1.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 3.7 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 5.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 3.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 6.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 5.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 1.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 4.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 3.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |