Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Onecut2_Onecut3
Z-value: 0.92
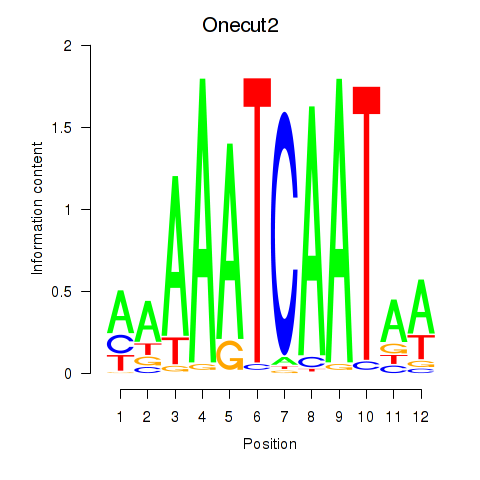
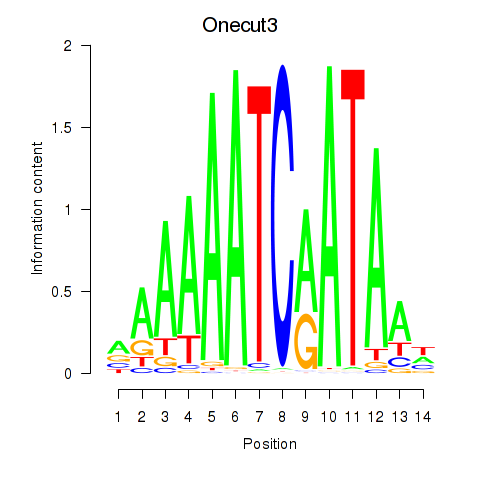
Transcription factors associated with Onecut2_Onecut3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Onecut2
|
ENSMUSG00000045991.20 | Onecut2 |
|
Onecut3
|
ENSMUSG00000045518.9 | Onecut3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Onecut3 | mm39_v1_chr10_+_80330669_80330714 | -0.43 | 8.3e-03 | Click! |
| Onecut2 | mm39_v1_chr18_+_64473091_64473114 | 0.36 | 3.1e-02 | Click! |
Activity profile of Onecut2_Onecut3 motif
Sorted Z-values of Onecut2_Onecut3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Onecut2_Onecut3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_61972348 | 4.79 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr9_-_86577940 | 4.75 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr6_-_128503666 | 4.45 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr3_-_81883509 | 4.37 |
ENSMUST00000029645.14
ENSMUST00000193879.2 |
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr15_+_10224052 | 4.36 |
ENSMUST00000128450.8
ENSMUST00000148257.8 ENSMUST00000128921.8 |
Prlr
|
prolactin receptor |
| chr3_+_20011251 | 2.86 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr3_+_20011201 | 2.73 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr3_+_20011405 | 2.46 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr16_-_10360893 | 2.22 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr7_+_119360141 | 2.21 |
ENSMUST00000106528.8
ENSMUST00000106527.8 ENSMUST00000063770.10 ENSMUST00000106529.8 |
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr6_-_21852508 | 2.07 |
ENSMUST00000031678.10
|
Tspan12
|
tetraspanin 12 |
| chr7_+_43856724 | 2.06 |
ENSMUST00000077354.5
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr10_+_128089965 | 1.88 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr1_-_180083859 | 1.80 |
ENSMUST00000111108.10
|
Psen2
|
presenilin 2 |
| chr1_+_171041583 | 1.65 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_+_51537645 | 1.62 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr1_+_171041539 | 1.58 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr5_-_87288177 | 1.46 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr15_-_89361571 | 1.41 |
ENSMUST00000165199.8
|
Arsa
|
arylsulfatase A |
| chr11_+_78389913 | 1.40 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr6_-_85879510 | 1.32 |
ENSMUST00000159755.8
|
Nat8f4
|
N-acetyltransferase 8 (GCN5-related) family member 4 |
| chr7_-_27010068 | 1.27 |
ENSMUST00000125455.2
|
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chr19_-_30152814 | 1.26 |
ENSMUST00000025778.9
|
Gldc
|
glycine decarboxylase |
| chr15_+_54975713 | 1.26 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr9_+_55234197 | 1.09 |
ENSMUST00000085754.10
ENSMUST00000034862.5 |
Tmem266
|
transmembrane protein 266 |
| chr14_-_4488167 | 1.05 |
ENSMUST00000022304.12
|
Thrb
|
thyroid hormone receptor beta |
| chr1_+_88030951 | 1.02 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr8_-_118398264 | 1.01 |
ENSMUST00000037955.14
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr1_+_167426019 | 0.99 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr9_-_51874846 | 0.98 |
ENSMUST00000034552.8
ENSMUST00000214013.2 |
Fdx1
|
ferredoxin 1 |
| chr1_+_172525613 | 0.98 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr17_-_35081129 | 0.93 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr17_-_35081456 | 0.92 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr8_-_85500010 | 0.92 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr12_+_103498542 | 0.91 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr11_+_97576619 | 0.90 |
ENSMUST00000107584.8
ENSMUST00000107585.9 |
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr15_-_96929086 | 0.89 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr2_-_147888816 | 0.86 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr17_-_32639936 | 0.81 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr1_+_34199333 | 0.81 |
ENSMUST00000183302.6
ENSMUST00000185897.7 ENSMUST00000185269.7 |
Dst
|
dystonin |
| chr18_+_20380397 | 0.81 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr2_-_91025441 | 0.79 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr15_+_54975814 | 0.79 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr10_+_128626772 | 0.77 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr13_+_60749735 | 0.77 |
ENSMUST00000226059.2
ENSMUST00000077453.13 |
Dapk1
|
death associated protein kinase 1 |
| chr7_+_130633776 | 0.73 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr11_+_97576724 | 0.73 |
ENSMUST00000107583.3
|
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr5_-_147831610 | 0.72 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr8_-_85500998 | 0.66 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr1_+_167425953 | 0.65 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr7_-_49286594 | 0.60 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr2_-_91025492 | 0.60 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr10_-_89568106 | 0.58 |
ENSMUST00000020109.5
|
Actr6
|
ARP6 actin-related protein 6 |
| chr3_-_10416369 | 0.57 |
ENSMUST00000108377.8
ENSMUST00000037839.12 |
Zfand1
|
zinc finger, AN1-type domain 1 |
| chr12_+_59178072 | 0.57 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr4_+_135648041 | 0.55 |
ENSMUST00000030434.5
|
Fuca1
|
fucosidase, alpha-L- 1, tissue |
| chr2_-_93988229 | 0.54 |
ENSMUST00000028619.5
|
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr13_+_60749995 | 0.54 |
ENSMUST00000044083.9
|
Dapk1
|
death associated protein kinase 1 |
| chr10_-_125225298 | 0.53 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr2_-_94236991 | 0.53 |
ENSMUST00000111237.9
ENSMUST00000094801.5 ENSMUST00000111238.8 |
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr6_+_48906825 | 0.52 |
ENSMUST00000031837.8
|
Doxl1
|
diamine oxidase-like protein 1 |
| chr13_+_119625345 | 0.52 |
ENSMUST00000099147.5
|
Tmem267
|
transmembrane protein 267 |
| chr7_+_112118817 | 0.50 |
ENSMUST00000106640.2
|
Parva
|
parvin, alpha |
| chr12_+_59178258 | 0.49 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr7_-_34354924 | 0.49 |
ENSMUST00000032709.3
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr8_-_22966831 | 0.48 |
ENSMUST00000163774.3
ENSMUST00000033935.16 |
Smim19
|
small integral membrane protein 19 |
| chr4_-_109013807 | 0.48 |
ENSMUST00000161363.2
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr7_-_90125178 | 0.48 |
ENSMUST00000032843.9
|
Tmem126b
|
transmembrane protein 126B |
| chr2_-_10135449 | 0.47 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr7_+_30676465 | 0.46 |
ENSMUST00000058093.6
|
Fam187b
|
family with sequence similarity 187, member B |
| chr2_+_144435974 | 0.46 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr1_-_136881336 | 0.46 |
ENSMUST00000192929.6
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr2_-_160169414 | 0.45 |
ENSMUST00000099127.3
|
Gm826
|
predicted gene 826 |
| chr17_+_45817750 | 0.44 |
ENSMUST00000024733.9
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr3_+_137849608 | 0.44 |
ENSMUST00000159481.8
ENSMUST00000161141.2 |
Trmt10a
|
tRNA methyltransferase 10A |
| chr19_+_44980565 | 0.43 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr18_-_24663260 | 0.42 |
ENSMUST00000046206.5
|
Rprd1a
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr15_+_92495007 | 0.42 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr9_-_64160899 | 0.42 |
ENSMUST00000005066.9
|
Map2k1
|
mitogen-activated protein kinase kinase 1 |
| chrX_-_112095181 | 0.41 |
ENSMUST00000026607.15
ENSMUST00000113388.3 |
Chm
|
choroidermia (RAB escort protein 1) |
| chr8_+_48277493 | 0.41 |
ENSMUST00000038693.8
|
Cldn22
|
claudin 22 |
| chr14_-_20231871 | 0.40 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr8_+_123103348 | 0.39 |
ENSMUST00000017622.12
ENSMUST00000093073.12 ENSMUST00000176699.2 |
Zc3h18
|
zinc finger CCCH-type containing 18 |
| chr2_-_63014622 | 0.39 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr6_-_67743756 | 0.38 |
ENSMUST00000103306.2
|
Igkv1-131
|
immunoglobulin kappa variable 1-131 |
| chr18_+_36431732 | 0.37 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chrX_-_110446022 | 0.37 |
ENSMUST00000156639.2
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr14_-_56448874 | 0.37 |
ENSMUST00000022757.5
|
Gzmf
|
granzyme F |
| chr6_-_52160816 | 0.35 |
ENSMUST00000134831.2
|
Hoxa3
|
homeobox A3 |
| chr16_-_28748410 | 0.35 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr18_+_77877611 | 0.35 |
ENSMUST00000238172.2
|
Pstpip2
|
proline-serine-threonine phosphatase-interacting protein 2 |
| chr13_-_22913799 | 0.34 |
ENSMUST00000237024.2
ENSMUST00000236800.2 |
Vmn1r207
|
vomeronasal 1 receptor 207 |
| chr7_+_18853778 | 0.32 |
ENSMUST00000053109.5
|
Fbxo46
|
F-box protein 46 |
| chr1_+_138201460 | 0.31 |
ENSMUST00000027643.6
|
Atp6v1g3
|
ATPase, H+ transporting, lysosomal V1 subunit G3 |
| chr8_-_73188887 | 0.31 |
ENSMUST00000109974.2
|
Calr3
|
calreticulin 3 |
| chr17_+_79359617 | 0.31 |
ENSMUST00000233916.2
|
Qpct
|
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
| chr17_-_35023521 | 0.31 |
ENSMUST00000025223.9
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr9_+_21279802 | 0.30 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chrX_+_106193060 | 0.30 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr6_-_92458324 | 0.29 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr3_-_98496123 | 0.29 |
ENSMUST00000178221.4
|
Gm10681
|
predicted gene 10681 |
| chr9_+_57604895 | 0.29 |
ENSMUST00000034865.6
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr6_-_127746390 | 0.29 |
ENSMUST00000032500.9
|
Prmt8
|
protein arginine N-methyltransferase 8 |
| chr4_+_145237329 | 0.29 |
ENSMUST00000105742.8
ENSMUST00000136309.8 |
Zfp990
|
zinc finger protein 990 |
| chr12_+_76812301 | 0.28 |
ENSMUST00000041262.14
ENSMUST00000126408.2 ENSMUST00000110399.3 ENSMUST00000137826.8 |
Churc1
Fntb
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta |
| chr6_+_127049865 | 0.28 |
ENSMUST00000000186.9
|
Fgf23
|
fibroblast growth factor 23 |
| chr17_+_35327255 | 0.28 |
ENSMUST00000037849.3
|
Ly6g5c
|
lymphocyte antigen 6 complex, locus G5C |
| chr2_-_63014514 | 0.27 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr13_-_22961570 | 0.27 |
ENSMUST00000227136.2
|
Vmn1r208
|
vomeronasal 1 receptor 208 |
| chr19_+_38253077 | 0.26 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr11_-_99241924 | 0.26 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
| chr8_+_22966736 | 0.25 |
ENSMUST00000067786.9
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr4_+_146033882 | 0.25 |
ENSMUST00000105730.2
ENSMUST00000091878.6 |
Zfp987
|
zinc finger protein 987 |
| chr19_+_38253105 | 0.24 |
ENSMUST00000196090.2
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr7_+_108549545 | 0.24 |
ENSMUST00000207583.2
|
Tub
|
tubby bipartite transcription factor |
| chr2_-_85592444 | 0.24 |
ENSMUST00000214255.2
|
Olfr1012
|
olfactory receptor 1012 |
| chr10_-_107321938 | 0.24 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr6_+_78382131 | 0.24 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr8_+_22966889 | 0.24 |
ENSMUST00000209305.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr3_+_115801869 | 0.24 |
ENSMUST00000106502.2
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr12_-_88288251 | 0.23 |
ENSMUST00000177747.2
|
Eif1ad15
|
eukaryotic translation initiation factor 1A domain containing 15 |
| chr2_+_81883566 | 0.23 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr7_+_101512922 | 0.23 |
ENSMUST00000209334.2
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr8_-_68270936 | 0.22 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_+_88640042 | 0.22 |
ENSMUST00000181601.2
|
Gm26566
|
predicted gene, 26566 |
| chrX_+_106193167 | 0.22 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chrX_+_118836893 | 0.21 |
ENSMUST00000040961.3
ENSMUST00000113366.2 |
Pabpc5
|
poly(A) binding protein, cytoplasmic 5 |
| chr8_-_88686188 | 0.21 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr11_+_87590720 | 0.21 |
ENSMUST00000040089.5
|
Rnf43
|
ring finger protein 43 |
| chr7_+_123061535 | 0.21 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr8_+_67149815 | 0.20 |
ENSMUST00000212588.2
|
Npy1r
|
neuropeptide Y receptor Y1 |
| chr13_+_35059285 | 0.20 |
ENSMUST00000077853.5
|
Prpf4b
|
pre-mRNA processing factor 4B |
| chr13_+_67052978 | 0.20 |
ENSMUST00000168767.9
|
Gm10767
|
predicted gene 10767 |
| chr7_-_23907518 | 0.19 |
ENSMUST00000086006.12
|
Zfp111
|
zinc finger protein 111 |
| chr14_-_68893253 | 0.19 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr4_+_146093394 | 0.19 |
ENSMUST00000168483.9
|
Zfp600
|
zinc finger protein 600 |
| chr13_-_55563028 | 0.19 |
ENSMUST00000054146.5
|
Pfn3
|
profilin 3 |
| chr8_-_72124359 | 0.19 |
ENSMUST00000177517.8
ENSMUST00000030170.15 |
Unc13a
|
unc-13 homolog A |
| chr6_-_42301488 | 0.19 |
ENSMUST00000095974.4
|
Fam131b
|
family with sequence similarity 131, member B |
| chr2_+_36308959 | 0.19 |
ENSMUST00000216645.2
|
Olfr339
|
olfactory receptor 339 |
| chr4_+_147106307 | 0.18 |
ENSMUST00000075775.6
|
Rex2
|
reduced expression 2 |
| chr12_-_36268171 | 0.18 |
ENSMUST00000020853.8
|
Lrrc72
|
leucine rich repeat containing 72 |
| chr18_+_62790356 | 0.18 |
ENSMUST00000162511.3
|
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr15_+_102927366 | 0.18 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr6_+_134617903 | 0.18 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr17_-_90763300 | 0.17 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr16_-_10131804 | 0.17 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2 |
| chr16_+_48130819 | 0.16 |
ENSMUST00000097175.5
|
Dppa2
|
developmental pluripotency associated 2 |
| chr9_+_78522783 | 0.16 |
ENSMUST00000093812.5
|
Cd109
|
CD109 antigen |
| chr13_-_22993852 | 0.16 |
ENSMUST00000227038.2
ENSMUST00000227265.2 |
Vmn1r209
|
vomeronasal 1 receptor 209 |
| chr1_-_5140504 | 0.16 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr13_-_8998267 | 0.16 |
ENSMUST00000187196.7
|
Gm9745
|
predicted gene 9745 |
| chr5_+_31684331 | 0.16 |
ENSMUST00000114533.9
ENSMUST00000202214.4 ENSMUST00000201858.4 ENSMUST00000202950.4 |
Slc4a1ap
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr12_+_4819022 | 0.16 |
ENSMUST00000219503.2
|
Pfn4
|
profilin family, member 4 |
| chr3_-_30194559 | 0.16 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr2_+_111156865 | 0.16 |
ENSMUST00000208176.3
|
Olfr1281
|
olfactory receptor 1281 |
| chr13_+_9002896 | 0.16 |
ENSMUST00000038598.3
|
Idi2
|
isopentenyl-diphosphate delta isomerase 2 |
| chr6_+_116185077 | 0.16 |
ENSMUST00000204051.2
|
Washc2
|
WASH complex subunit 2` |
| chr2_-_111942878 | 0.15 |
ENSMUST00000217078.2
|
Olfr1315-ps1
|
olfactory receptor 1315, pseudogene 1 |
| chr11_+_115197980 | 0.15 |
ENSMUST00000055490.9
|
Otop2
|
otopetrin 2 |
| chr6_+_141470105 | 0.15 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr17_-_24199523 | 0.15 |
ENSMUST00000234765.2
|
Prss30
|
protease, serine 30 |
| chr18_+_37646674 | 0.15 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr8_-_5155347 | 0.15 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr9_+_45749869 | 0.15 |
ENSMUST00000078111.11
ENSMUST00000034591.11 |
Bace1
|
beta-site APP cleaving enzyme 1 |
| chr6_+_116184991 | 0.14 |
ENSMUST00000036759.11
ENSMUST00000204476.3 |
Washc2
|
WASH complex subunit 2` |
| chr7_-_119461027 | 0.14 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr7_+_123061497 | 0.14 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr4_+_43384320 | 0.14 |
ENSMUST00000136360.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr6_+_116483477 | 0.14 |
ENSMUST00000075756.3
|
Olfr212
|
olfactory receptor 212 |
| chr6_-_122833109 | 0.14 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1 |
| chr5_-_52723700 | 0.14 |
ENSMUST00000039750.7
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr1_-_170042947 | 0.13 |
ENSMUST00000027979.14
ENSMUST00000123399.2 |
Uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr5_-_74838461 | 0.13 |
ENSMUST00000117525.8
ENSMUST00000113531.9 ENSMUST00000039744.13 ENSMUST00000121690.8 |
Lnx1
|
ligand of numb-protein X 1 |
| chr10_+_76089674 | 0.13 |
ENSMUST00000036387.8
|
S100b
|
S100 protein, beta polypeptide, neural |
| chr19_+_38252984 | 0.13 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr7_+_87233554 | 0.13 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chr10_+_77885669 | 0.13 |
ENSMUST00000000746.12
|
Dnmt3l
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr7_-_37722938 | 0.13 |
ENSMUST00000206581.2
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr3_+_91921890 | 0.13 |
ENSMUST00000047660.5
|
Pglyrp3
|
peptidoglycan recognition protein 3 |
| chrX_+_105059305 | 0.12 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr2_+_59442378 | 0.12 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chrX_+_106192510 | 0.12 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr2_-_30249202 | 0.12 |
ENSMUST00000100215.11
ENSMUST00000113620.10 ENSMUST00000163668.3 ENSMUST00000028214.15 ENSMUST00000113621.10 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr2_-_17395765 | 0.12 |
ENSMUST00000177966.2
|
Nebl
|
nebulette |
| chr3_+_29136172 | 0.12 |
ENSMUST00000124809.8
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr7_-_140087224 | 0.12 |
ENSMUST00000209873.2
ENSMUST00000064392.8 ENSMUST00000215768.2 ENSMUST00000215340.2 |
Olfr536
|
olfactory receptor 536 |
| chr16_+_95059121 | 0.12 |
ENSMUST00000113858.3
|
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr7_+_27290969 | 0.12 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr14_-_50536787 | 0.12 |
ENSMUST00000163469.2
|
Olfr733
|
olfactory receptor 733 |
| chr7_-_103928939 | 0.12 |
ENSMUST00000051795.10
|
Trim5
|
tripartite motif-containing 5 |
| chr10_+_126836578 | 0.11 |
ENSMUST00000026500.12
ENSMUST00000142698.8 |
Avil
|
advillin |
| chrY_+_897782 | 0.11 |
ENSMUST00000189069.7
ENSMUST00000055032.14 |
Kdm5d
|
lysine (K)-specific demethylase 5D |
| chr16_-_45352346 | 0.11 |
ENSMUST00000232600.2
|
Gm17783
|
predicted gene, 17783 |
| chr17_+_43671314 | 0.11 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr4_-_115797118 | 0.11 |
ENSMUST00000124071.9
ENSMUST00000084338.7 |
Dmbx1
|
diencephalon/mesencephalon homeobox 1 |
| chr13_-_103042554 | 0.11 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_-_98220466 | 0.11 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr4_+_147576874 | 0.11 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chrX_-_74205226 | 0.11 |
ENSMUST00000165080.2
|
Smim9
|
small integral membrane protein 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.8 | 4.8 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.7 | 4.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.6 | 4.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.4 | 1.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 1.4 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.3 | 1.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.3 | 0.3 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.3 | 0.9 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 0.9 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.2 | 8.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 1.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 0.9 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 1.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.9 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 1.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 2.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.5 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 4.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.4 | GO:0010159 | specification of organ position(GO:0010159) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 1.0 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 2.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 2.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.4 | GO:1903800 | regulation of Golgi inheritance(GO:0090170) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 1.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0097473 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.5 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.3 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.6 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 1.4 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 2.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.1 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.0 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.3 | GO:0042448 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) progesterone metabolic process(GO:0042448) |
| 0.0 | 0.3 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.5 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0001967 | suckling behavior(GO:0001967) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 8.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.3 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 1.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.9 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.1 | 4.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.0 | 4.8 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.9 | 4.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.7 | 7.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 4.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.3 | 4.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 2.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 1.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.2 | 1.4 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 0.5 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 1.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 1.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.9 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.3 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 2.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 1.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 1.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 1.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 2.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.7 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 3.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 8.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 2.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 1.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.8 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 7.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 4.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |