Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
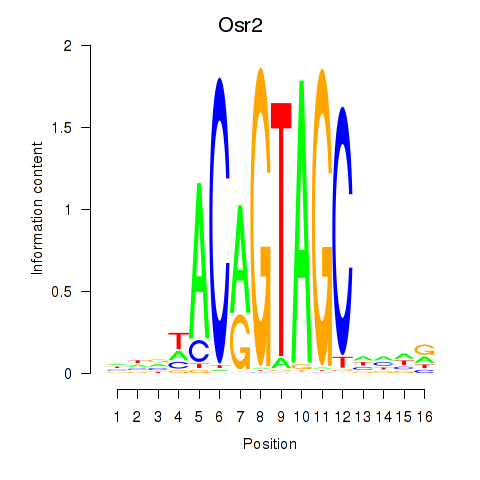
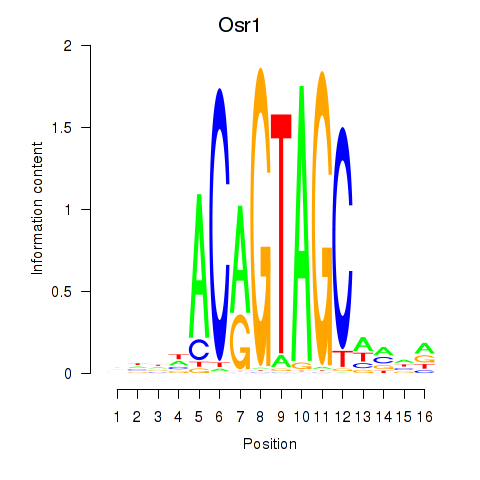
Results for Osr2_Osr1
Z-value: 0.47
Transcription factors associated with Osr2_Osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Osr2
|
ENSMUSG00000022330.6 | Osr2 |
|
Osr1
|
ENSMUSG00000048387.9 | Osr1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Osr1 | mm39_v1_chr12_+_9624437_9624448 | 0.15 | 4.0e-01 | Click! |
| Osr2 | mm39_v1_chr15_+_35296237_35296250 | 0.12 | 5.0e-01 | Click! |
Activity profile of Osr2_Osr1 motif
Sorted Z-values of Osr2_Osr1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Osr2_Osr1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_98670369 | 2.64 |
ENSMUST00000107019.8
ENSMUST00000107018.8 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr17_-_57535003 | 2.45 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr1_+_88066086 | 2.00 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr19_+_20579322 | 1.83 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr1_-_180823709 | 1.41 |
ENSMUST00000154133.8
|
Ephx1
|
epoxide hydrolase 1, microsomal |
| chr5_-_89583469 | 1.18 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr4_-_115504907 | 1.11 |
ENSMUST00000102707.10
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr3_+_118355778 | 1.08 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr13_+_55547498 | 1.06 |
ENSMUST00000057167.9
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr3_+_129630380 | 1.00 |
ENSMUST00000077918.7
|
Cfi
|
complement component factor i |
| chr16_-_18232202 | 0.89 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr3_-_98669720 | 0.78 |
ENSMUST00000146196.4
|
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr5_-_87240405 | 0.77 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr18_+_60515755 | 0.66 |
ENSMUST00000237185.2
|
Iigp1
|
interferon inducible GTPase 1 |
| chr18_+_21077627 | 0.63 |
ENSMUST00000050004.3
|
Rnf125
|
ring finger protein 125 |
| chr9_+_48406706 | 0.62 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr6_+_85888850 | 0.57 |
ENSMUST00000200680.4
|
Tprkb
|
Tp53rk binding protein |
| chr13_-_4329421 | 0.55 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr9_+_108539296 | 0.54 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr7_+_44499374 | 0.53 |
ENSMUST00000141311.8
ENSMUST00000107880.9 ENSMUST00000208384.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr13_-_74498320 | 0.52 |
ENSMUST00000221594.2
ENSMUST00000022062.8 |
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr4_+_139350152 | 0.51 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr14_+_77274185 | 0.51 |
ENSMUST00000048208.10
ENSMUST00000095625.11 |
Ccdc122
|
coiled-coil domain containing 122 |
| chr15_+_81686622 | 0.50 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr19_+_18609291 | 0.49 |
ENSMUST00000042392.14
ENSMUST00000237347.2 |
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr2_+_34764408 | 0.47 |
ENSMUST00000113068.9
ENSMUST00000047447.13 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr3_+_118355811 | 0.46 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr17_-_28779678 | 0.46 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr17_+_56469477 | 0.45 |
ENSMUST00000077788.7
|
Tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr15_-_89314054 | 0.45 |
ENSMUST00000023289.13
|
Chkb
|
choline kinase beta |
| chr9_-_110571645 | 0.44 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr2_-_86180622 | 0.44 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr12_+_113106407 | 0.44 |
ENSMUST00000196015.5
|
Crip2
|
cysteine rich protein 2 |
| chr19_+_37423198 | 0.43 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr10_+_43777777 | 0.42 |
ENSMUST00000054418.12
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr2_-_177567397 | 0.41 |
ENSMUST00000108934.9
ENSMUST00000081529.11 |
Zfp972
|
zinc finger protein 972 |
| chr6_-_142517340 | 0.40 |
ENSMUST00000203945.3
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr10_-_39775182 | 0.39 |
ENSMUST00000178045.9
ENSMUST00000178563.3 |
Mfsd4b4
|
major facilitator superfamily domain containing 4B4 |
| chr12_+_85157607 | 0.38 |
ENSMUST00000053811.10
|
Dlst
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr2_+_34764496 | 0.38 |
ENSMUST00000028228.6
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr13_+_4241149 | 0.37 |
ENSMUST00000021634.4
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr2_-_5719302 | 0.37 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr12_-_113393106 | 0.36 |
ENSMUST00000192746.2
ENSMUST00000103429.2 |
Ighj2
|
immunoglobulin heavy joining 2 |
| chr2_-_164063525 | 0.36 |
ENSMUST00000018355.11
ENSMUST00000109376.9 |
Wfdc15b
|
WAP four-disulfide core domain 15B |
| chr10_-_23985432 | 0.36 |
ENSMUST00000041180.7
|
Taar9
|
trace amine-associated receptor 9 |
| chr7_+_119160922 | 0.35 |
ENSMUST00000130583.2
ENSMUST00000084647.13 |
Acsm2
|
acyl-CoA synthetase medium-chain family member 2 |
| chr6_-_142517029 | 0.34 |
ENSMUST00000032374.9
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr9_-_51240201 | 0.33 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr5_-_66238313 | 0.33 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr7_-_127490139 | 0.33 |
ENSMUST00000205300.2
ENSMUST00000121394.3 |
Prss53
|
protease, serine 53 |
| chr10_+_4561974 | 0.32 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr10_+_120044650 | 0.32 |
ENSMUST00000020446.11
ENSMUST00000134797.8 |
Tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr16_+_29884153 | 0.31 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr17_-_45904540 | 0.30 |
ENSMUST00000163905.8
ENSMUST00000167692.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr10_+_3690348 | 0.30 |
ENSMUST00000120274.8
|
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr6_-_59001455 | 0.29 |
ENSMUST00000089860.12
|
Fam13a
|
family with sequence similarity 13, member A |
| chr8_+_67933573 | 0.29 |
ENSMUST00000212171.2
|
Nat1
|
N-acetyl transferase 1 |
| chr19_+_18609343 | 0.29 |
ENSMUST00000159572.9
|
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr7_-_142253247 | 0.28 |
ENSMUST00000105934.8
|
Ins2
|
insulin II |
| chrX_+_138464065 | 0.28 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr10_+_84674008 | 0.28 |
ENSMUST00000095388.5
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr4_-_119240885 | 0.27 |
ENSMUST00000238422.2
ENSMUST00000238719.2 ENSMUST00000238723.2 ENSMUST00000044781.9 ENSMUST00000084307.5 ENSMUST00000148236.9 ENSMUST00000127474.3 ENSMUST00000238704.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr3_-_92393193 | 0.27 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr4_+_132948121 | 0.27 |
ENSMUST00000105910.2
|
Cd164l2
|
CD164 sialomucin-like 2 |
| chr19_+_5927876 | 0.27 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr7_+_44499005 | 0.25 |
ENSMUST00000150335.2
ENSMUST00000107882.8 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr8_-_94739469 | 0.24 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr9_+_45817795 | 0.24 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chrM_+_8603 | 0.23 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr7_-_123099672 | 0.23 |
ENSMUST00000042470.14
ENSMUST00000128217.2 |
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr6_+_51500881 | 0.22 |
ENSMUST00000049152.15
|
Snx10
|
sorting nexin 10 |
| chr7_-_37469049 | 0.21 |
ENSMUST00000175941.8
|
Zfp536
|
zinc finger protein 536 |
| chr16_+_16888145 | 0.21 |
ENSMUST00000232574.2
|
Ypel1
|
yippee like 1 |
| chr8_+_70583971 | 0.20 |
ENSMUST00000211898.2
ENSMUST00000095273.7 |
Nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr7_+_44498640 | 0.20 |
ENSMUST00000054343.15
ENSMUST00000142880.3 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr12_+_52144511 | 0.20 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr7_+_141503719 | 0.19 |
ENSMUST00000105989.9
ENSMUST00000075528.12 ENSMUST00000174499.8 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr11_+_72192455 | 0.19 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr15_-_103446354 | 0.19 |
ENSMUST00000023133.8
|
Ppp1r1a
|
protein phosphatase 1, regulatory inhibitor subunit 1A |
| chr9_-_48406564 | 0.19 |
ENSMUST00000213276.2
ENSMUST00000170000.4 |
Rbm7
|
RNA binding motif protein 7 |
| chr15_+_78819378 | 0.19 |
ENSMUST00000145157.2
ENSMUST00000123013.2 |
Nol12
|
nucleolar protein 12 |
| chr8_+_3671599 | 0.18 |
ENSMUST00000207389.2
|
Pet100
|
PET100 homolog |
| chr4_-_138641225 | 0.18 |
ENSMUST00000097830.4
|
Otud3
|
OTU domain containing 3 |
| chr14_-_51493569 | 0.18 |
ENSMUST00000061936.8
|
Rnase2a
|
ribonuclease, RNase A family, 2A (liver, eosinophil-derived neurotoxin) |
| chr1_+_74640590 | 0.18 |
ENSMUST00000087183.11
ENSMUST00000148456.8 ENSMUST00000113694.8 |
Stk36
|
serine/threonine kinase 36 |
| chr1_+_44159106 | 0.18 |
ENSMUST00000114709.3
ENSMUST00000129068.2 |
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr3_-_89230190 | 0.18 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr2_-_79959178 | 0.17 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_-_127292636 | 0.17 |
ENSMUST00000189368.2
|
Ank2
|
ankyrin 2, brain |
| chr7_+_141503411 | 0.17 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr9_+_45818250 | 0.17 |
ENSMUST00000216672.2
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr11_+_67689094 | 0.17 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr1_+_74640706 | 0.17 |
ENSMUST00000087186.11
|
Stk36
|
serine/threonine kinase 36 |
| chr6_+_67586695 | 0.17 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr10_+_78261503 | 0.17 |
ENSMUST00000005185.8
|
Cstb
|
cystatin B |
| chr6_-_59001325 | 0.17 |
ENSMUST00000173193.2
|
Fam13a
|
family with sequence similarity 13, member A |
| chr6_+_90346547 | 0.17 |
ENSMUST00000045740.8
ENSMUST00000203493.2 |
Zxdc
|
ZXD family zinc finger C |
| chr3_+_63148887 | 0.16 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr15_+_35371644 | 0.16 |
ENSMUST00000227455.2
|
Vps13b
|
vacuolar protein sorting 13B |
| chr14_-_31157985 | 0.16 |
ENSMUST00000091903.5
|
Sh3bp5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr5_+_106024398 | 0.15 |
ENSMUST00000150440.8
ENSMUST00000031227.11 |
Zfp326
|
zinc finger protein 326 |
| chr19_-_41195213 | 0.15 |
ENSMUST00000169941.2
ENSMUST00000025986.15 |
Tll2
|
tolloid-like 2 |
| chr1_-_93233572 | 0.14 |
ENSMUST00000112944.8
ENSMUST00000112942.2 ENSMUST00000027492.14 |
Mterf4
|
mitochondrial transcription termination factor 4 |
| chr12_+_112645237 | 0.13 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr19_-_18609118 | 0.13 |
ENSMUST00000025631.7
ENSMUST00000236615.2 |
Ostf1
|
osteoclast stimulating factor 1 |
| chr4_+_45890303 | 0.13 |
ENSMUST00000178561.8
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr8_+_46428551 | 0.13 |
ENSMUST00000034051.7
ENSMUST00000150943.2 |
Ufsp2
|
UFM1-specific peptidase 2 |
| chr17_-_7639378 | 0.13 |
ENSMUST00000231397.2
|
Gm49630
|
predicted gene, 49630 |
| chr10_-_128640232 | 0.13 |
ENSMUST00000051011.14
|
Tmem198b
|
transmembrane protein 198b |
| chr7_+_141503583 | 0.13 |
ENSMUST00000172652.8
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr5_+_114991722 | 0.13 |
ENSMUST00000031547.12
|
4930519G04Rik
|
RIKEN cDNA 4930519G04 gene |
| chr6_+_70700207 | 0.13 |
ENSMUST00000103407.3
ENSMUST00000199487.2 |
Igkj3
|
immunoglobulin kappa joining 3 |
| chr9_-_53521585 | 0.13 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr14_-_24054352 | 0.13 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_+_90346468 | 0.12 |
ENSMUST00000113539.8
ENSMUST00000075117.10 |
Zxdc
|
ZXD family zinc finger C |
| chr2_+_179713586 | 0.12 |
ENSMUST00000108901.8
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr9_-_55190902 | 0.12 |
ENSMUST00000164721.8
|
Nrg4
|
neuregulin 4 |
| chr8_+_3671528 | 0.12 |
ENSMUST00000156380.4
|
Pet100
|
PET100 homolog |
| chr11_-_70130620 | 0.12 |
ENSMUST00000040428.4
|
Rnasek
|
ribonuclease, RNase K |
| chr17_+_35327255 | 0.11 |
ENSMUST00000037849.3
|
Ly6g5c
|
lymphocyte antigen 6 complex, locus G5C |
| chr7_-_119319965 | 0.11 |
ENSMUST00000033236.9
|
Thumpd1
|
THUMP domain containing 1 |
| chr11_-_105347500 | 0.11 |
ENSMUST00000049995.10
ENSMUST00000100332.4 |
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr8_-_46428277 | 0.11 |
ENSMUST00000095323.8
ENSMUST00000098786.3 |
1700029J07Rik
|
RIKEN cDNA 1700029J07 gene |
| chr4_+_101944740 | 0.11 |
ENSMUST00000106911.8
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chrX_+_106711562 | 0.11 |
ENSMUST00000168174.9
|
Tbx22
|
T-box 22 |
| chr1_+_107289659 | 0.10 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr6_-_144155197 | 0.10 |
ENSMUST00000038815.14
ENSMUST00000111749.8 ENSMUST00000170367.9 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr13_+_92442488 | 0.10 |
ENSMUST00000188317.2
|
Gm20379
|
predicted gene, 20379 |
| chr11_+_8998575 | 0.10 |
ENSMUST00000043285.5
|
Gm11992
|
predicted gene 11992 |
| chr11_-_98040377 | 0.10 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr11_-_100650566 | 0.10 |
ENSMUST00000107361.9
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr7_+_126549859 | 0.09 |
ENSMUST00000106333.8
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr1_-_156248716 | 0.09 |
ENSMUST00000178036.8
|
Axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr10_-_129948657 | 0.09 |
ENSMUST00000081469.2
|
Olfr823
|
olfactory receptor 823 |
| chr9_-_16289527 | 0.09 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr3_-_19682833 | 0.09 |
ENSMUST00000119133.2
|
1700064H15Rik
|
RIKEN cDNA 1700064H15 gene |
| chr4_+_125997788 | 0.09 |
ENSMUST00000139524.2
|
Stk40
|
serine/threonine kinase 40 |
| chr12_+_84161095 | 0.09 |
ENSMUST00000123491.8
ENSMUST00000046340.9 ENSMUST00000136159.2 |
Dnal1
|
dynein, axonemal, light chain 1 |
| chr6_+_30512285 | 0.08 |
ENSMUST00000031798.14
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr7_+_96600712 | 0.08 |
ENSMUST00000044466.12
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr4_-_43771009 | 0.08 |
ENSMUST00000053931.2
|
Olfr159
|
olfactory receptor 159 |
| chr15_+_74388044 | 0.08 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr1_-_74640504 | 0.08 |
ENSMUST00000136078.2
ENSMUST00000132081.2 ENSMUST00000113721.8 ENSMUST00000027357.12 |
Rnf25
|
ring finger protein 25 |
| chr15_-_60793115 | 0.08 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chrX_+_162873183 | 0.08 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr12_-_111679344 | 0.08 |
ENSMUST00000160576.2
|
Bag5
|
BCL2-associated athanogene 5 |
| chr19_+_54033681 | 0.07 |
ENSMUST00000237285.2
|
Adra2a
|
adrenergic receptor, alpha 2a |
| chr2_-_103858632 | 0.07 |
ENSMUST00000056170.4
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr19_-_32080496 | 0.07 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr8_-_47805383 | 0.07 |
ENSMUST00000110367.10
|
Stox2
|
storkhead box 2 |
| chr8_+_27513819 | 0.07 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr12_-_110649040 | 0.07 |
ENSMUST00000222915.2
ENSMUST00000070659.7 |
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr7_-_57159743 | 0.07 |
ENSMUST00000068456.8
ENSMUST00000206734.2 |
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr6_-_86710250 | 0.07 |
ENSMUST00000001185.14
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr9_-_7184440 | 0.07 |
ENSMUST00000140466.8
|
Dync2h1
|
dynein cytoplasmic 2 heavy chain 1 |
| chr15_-_43733389 | 0.06 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr6_-_119940694 | 0.06 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr4_+_125997734 | 0.06 |
ENSMUST00000116286.9
ENSMUST00000094761.11 |
Stk40
|
serine/threonine kinase 40 |
| chr11_+_119804630 | 0.06 |
ENSMUST00000026434.13
ENSMUST00000124199.8 |
Chmp6
|
charged multivesicular body protein 6 |
| chr10_-_107321938 | 0.06 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr6_+_41512480 | 0.06 |
ENSMUST00000103289.2
ENSMUST00000103290.2 ENSMUST00000193061.2 |
Trbj1-6
Trbj1-7
|
T cell receptor beta joining 1-6 T cell receptor beta joining 1-7 |
| chrX_+_163219983 | 0.06 |
ENSMUST00000036858.11
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr2_-_69715899 | 0.06 |
ENSMUST00000060447.13
|
Mettl5
|
methyltransferase like 5 |
| chr6_-_69741999 | 0.06 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr6_+_67736650 | 0.06 |
ENSMUST00000103305.2
|
Igkv1-132
|
immunoglobulin kappa variable 1-132 |
| chr7_-_30443106 | 0.06 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr3_+_87343071 | 0.06 |
ENSMUST00000194102.6
|
Fcrl5
|
Fc receptor-like 5 |
| chr3_+_87343127 | 0.06 |
ENSMUST00000166297.7
ENSMUST00000049926.15 ENSMUST00000178261.3 |
Fcrl5
|
Fc receptor-like 5 |
| chr18_+_70058613 | 0.05 |
ENSMUST00000080050.6
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr8_-_25085654 | 0.05 |
ENSMUST00000110667.8
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chrY_-_10750826 | 0.05 |
ENSMUST00000177818.2
|
Gm20777
|
predicted gene, 20777 |
| chrX_+_52523795 | 0.05 |
ENSMUST00000074232.7
|
Etd
|
embryonic testis differentiation |
| chr3_+_87343105 | 0.05 |
ENSMUST00000193229.6
|
Fcrl5
|
Fc receptor-like 5 |
| chrY_-_10534428 | 0.05 |
ENSMUST00000179665.2
|
Gm20737
|
predicted gene, 20737 |
| chr2_+_103242027 | 0.05 |
ENSMUST00000239273.2
ENSMUST00000164172.8 |
Elf5
|
E74-like factor 5 |
| chr7_+_96600756 | 0.05 |
ENSMUST00000107159.3
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr4_-_24800890 | 0.05 |
ENSMUST00000108214.9
|
Klhl32
|
kelch-like 32 |
| chr3_-_144514386 | 0.05 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr12_+_71021395 | 0.05 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr1_+_106862171 | 0.05 |
ENSMUST00000081277.9
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr6_-_42574306 | 0.04 |
ENSMUST00000069023.4
|
Tcaf3
|
TRPM8 channel-associated factor 3 |
| chr6_+_141470261 | 0.04 |
ENSMUST00000203140.2
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr5_+_9316097 | 0.04 |
ENSMUST00000134991.8
ENSMUST00000069538.14 ENSMUST00000115348.9 |
Elapor2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr15_-_36140539 | 0.04 |
ENSMUST00000172831.8
|
Rgs22
|
regulator of G-protein signalling 22 |
| chr11_-_32150222 | 0.04 |
ENSMUST00000145401.8
ENSMUST00000142396.2 ENSMUST00000128311.8 |
Il9r
|
interleukin 9 receptor |
| chr10_-_80374916 | 0.04 |
ENSMUST00000219648.2
|
Atp8b3
|
ATPase, class I, type 8B, member 3 |
| chr11_+_51111300 | 0.04 |
ENSMUST00000126189.3
|
Msantd5
|
Myb/SANT DNA binding domain containing 5 |
| chr6_-_68907718 | 0.04 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr12_-_73093953 | 0.04 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr4_-_116413092 | 0.04 |
ENSMUST00000069674.6
ENSMUST00000106478.9 |
Tmem69
|
transmembrane protein 69 |
| chr9_+_75255037 | 0.04 |
ENSMUST00000034709.7
|
Bcl2l10
|
Bcl2-like 10 |
| chr1_-_85888729 | 0.04 |
ENSMUST00000086975.7
|
Gpr55
|
G protein-coupled receptor 55 |
| chr17_+_8454862 | 0.04 |
ENSMUST00000231340.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr14_+_55842002 | 0.04 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr9_-_45818196 | 0.04 |
ENSMUST00000160699.9
|
Rnf214
|
ring finger protein 214 |
| chr12_-_75224099 | 0.04 |
ENSMUST00000042299.4
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr15_-_27681643 | 0.04 |
ENSMUST00000100739.5
|
Otulinl
|
OTU deubiquitinase with linear linkage specificity like |
| chr7_+_45175754 | 0.04 |
ENSMUST00000211227.2
ENSMUST00000051810.15 |
Plekha4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr7_-_6511472 | 0.03 |
ENSMUST00000207055.3
ENSMUST00000209097.2 ENSMUST00000208623.3 ENSMUST00000208207.2 ENSMUST00000209029.4 ENSMUST00000214383.2 ENSMUST00000207624.2 ENSMUST00000213549.2 |
Olfr1348
|
olfactory receptor 1348 |
| chr9_-_45818134 | 0.03 |
ENSMUST00000161203.8
ENSMUST00000058720.13 |
Rnf214
|
ring finger protein 214 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.5 | 1.5 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.4 | 1.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.4 | 1.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 0.9 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.2 | 1.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 2.0 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0042668 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.1 | 0.3 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 1.4 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.1 | 0.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.4 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 0.5 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 1.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 1.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.8 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.4 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.0 | 0.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 3.4 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.9 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.4 | GO:0006103 | tricarboxylic acid cycle(GO:0006099) 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.5 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.2 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 3.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 3.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.5 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.5 | 3.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.4 | 1.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 0.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 1.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.3 | 1.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.3 | 0.8 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.2 | 1.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 0.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.5 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.4 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.3 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 3.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |