Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pax2
Z-value: 2.50
Transcription factors associated with Pax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax2
|
ENSMUSG00000004231.16 | Pax2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax2 | mm39_v1_chr19_+_44745833_44745833 | -0.24 | 1.7e-01 | Click! |
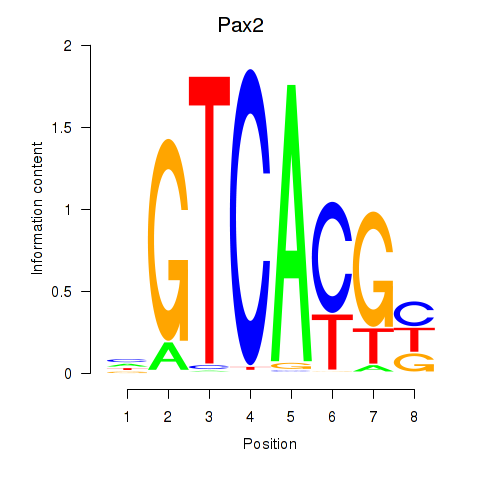
Activity profile of Pax2 motif
Sorted Z-values of Pax2 motif
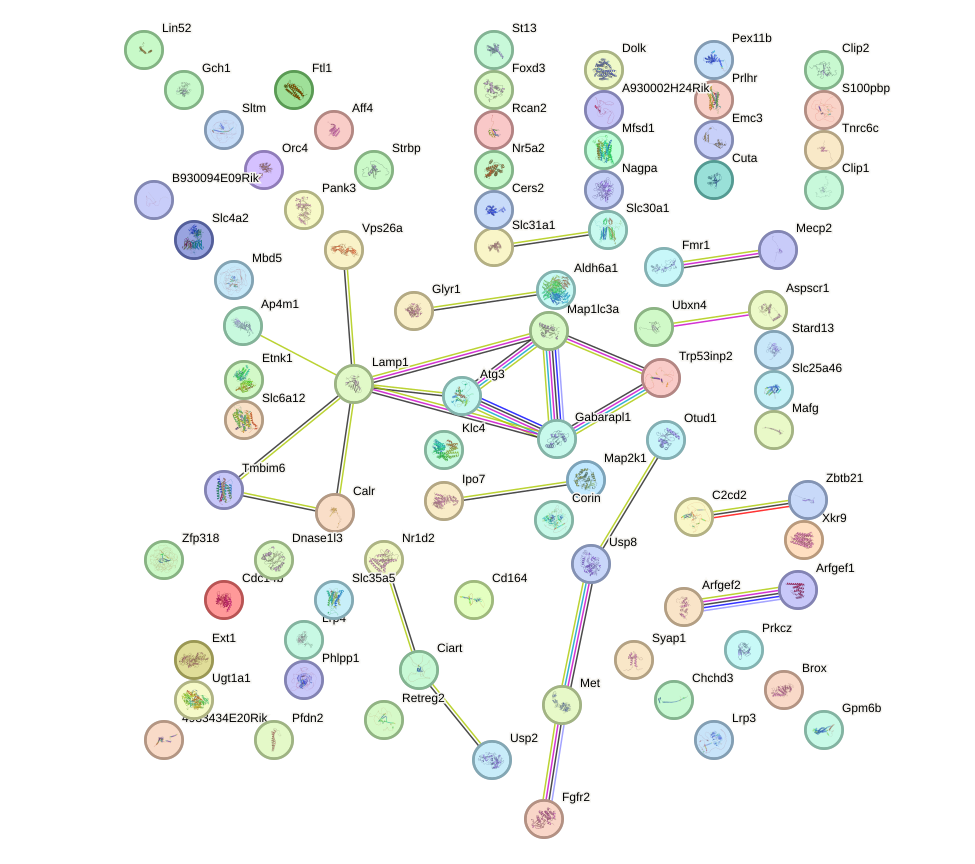
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_84497718 | 10.87 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr14_+_55798362 | 7.44 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr14_+_55797468 | 7.16 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr14_+_55797934 | 6.97 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr7_-_34914675 | 6.53 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr3_+_89960121 | 6.30 |
ENSMUST00000160640.8
ENSMUST00000029552.13 ENSMUST00000162114.8 ENSMUST00000068798.13 |
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr8_+_13209141 | 5.78 |
ENSMUST00000033824.8
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr16_-_97763780 | 5.54 |
ENSMUST00000232187.2
ENSMUST00000231263.2 ENSMUST00000052089.9 ENSMUST00000063605.15 ENSMUST00000113734.9 ENSMUST00000231560.2 ENSMUST00000232165.2 |
Zbtb21
C2cd2
|
zinc finger and BTB domain containing 21 C2 calcium-dependent domain containing 2 |
| chr16_+_44979086 | 5.49 |
ENSMUST00000023343.4
|
Atg3
|
autophagy related 3 |
| chr14_-_47426863 | 5.41 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr10_-_62322551 | 4.48 |
ENSMUST00000105447.11
|
Vps26a
|
VPS26 retromer complex component A |
| chr17_-_46956920 | 4.44 |
ENSMUST00000233974.2
|
Klc4
|
kinesin light chain 4 |
| chr17_-_27158514 | 4.29 |
ENSMUST00000114935.9
ENSMUST00000025027.10 |
Cuta
|
cutA divalent cation tolerance homolog |
| chr4_-_148123223 | 4.24 |
ENSMUST00000030879.12
ENSMUST00000137724.8 |
Clcn6
|
chloride channel, voltage-sensitive 6 |
| chr10_+_41395410 | 4.07 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr6_+_129510117 | 4.06 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr14_+_4230569 | 4.05 |
ENSMUST00000090543.6
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr1_+_128171859 | 3.97 |
ENSMUST00000027592.6
|
Ubxn4
|
UBX domain protein 4 |
| chr14_+_55797443 | 3.91 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr1_+_13738967 | 3.88 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr1_+_106099482 | 3.86 |
ENSMUST00000061047.7
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr6_+_17463925 | 3.82 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr16_-_44979013 | 3.72 |
ENSMUST00000023344.10
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr3_+_67490068 | 3.61 |
ENSMUST00000029344.10
|
Mfsd1
|
major facilitator superfamily domain containing 1 |
| chr14_+_14475188 | 3.54 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr3_+_95226093 | 3.52 |
ENSMUST00000139866.2
|
Cers2
|
ceramide synthase 2 |
| chr10_-_62322356 | 3.44 |
ENSMUST00000092473.5
|
Vps26a
|
VPS26 retromer complex component A |
| chr10_+_41395870 | 3.40 |
ENSMUST00000189300.2
|
Cd164
|
CD164 antigen |
| chr15_+_99291100 | 3.39 |
ENSMUST00000159209.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr15_+_99291491 | 3.37 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr15_-_81283795 | 3.36 |
ENSMUST00000023039.15
|
St13
|
suppression of tumorigenicity 13 |
| chr15_+_99291455 | 3.20 |
ENSMUST00000162624.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr3_-_146302343 | 3.09 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr4_-_155430153 | 3.08 |
ENSMUST00000103178.11
|
Prkcz
|
protein kinase C, zeta |
| chr1_+_88139678 | 2.97 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr9_-_64160899 | 2.97 |
ENSMUST00000005066.9
|
Map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr1_-_136888118 | 2.96 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr11_+_35660288 | 2.95 |
ENSMUST00000018990.8
|
Pank3
|
pantothenate kinase 3 |
| chr15_-_81284244 | 2.89 |
ENSMUST00000172107.8
ENSMUST00000169204.2 ENSMUST00000163382.2 |
St13
|
suppression of tumorigenicity 13 |
| chr16_-_44978929 | 2.87 |
ENSMUST00000181177.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr4_+_62278932 | 2.79 |
ENSMUST00000084526.12
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr5_-_151113619 | 2.78 |
ENSMUST00000062015.15
ENSMUST00000110483.9 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr10_-_62322451 | 2.76 |
ENSMUST00000217868.2
|
Vps26a
|
VPS26 retromer complex component A |
| chr13_-_64422775 | 2.76 |
ENSMUST00000221634.2
ENSMUST00000039318.16 |
Cdc14b
|
CDC14 cell division cycle 14B |
| chr2_+_164675697 | 2.76 |
ENSMUST00000143780.9
|
Ctsa
|
cathepsin A |
| chr1_+_75119419 | 2.74 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr1_-_183078488 | 2.70 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chrX_-_161671421 | 2.68 |
ENSMUST00000033723.4
|
Syap1
|
synapse associated protein 1 |
| chr16_-_44978986 | 2.67 |
ENSMUST00000180636.8
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr2_+_155223728 | 2.67 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr7_+_109617456 | 2.61 |
ENSMUST00000084731.5
|
Ipo7
|
importin 7 |
| chr1_+_75119472 | 2.60 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr1_-_175319842 | 2.59 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr17_-_64170786 | 2.58 |
ENSMUST00000050753.4
|
A930002H24Rik
|
RIKEN cDNA A930002H24 gene |
| chr6_-_113508536 | 2.56 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr18_-_31742946 | 2.55 |
ENSMUST00000060396.7
|
Slc25a46
|
solute carrier family 25, member 46 |
| chr6_+_90439596 | 2.55 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr6_+_90439544 | 2.53 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr4_-_129083251 | 2.52 |
ENSMUST00000117965.8
|
S100pbp
|
S100P binding protein |
| chr5_-_66309244 | 2.49 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr7_+_46445512 | 2.47 |
ENSMUST00000006774.11
ENSMUST00000165031.8 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr2_+_166647426 | 2.47 |
ENSMUST00000099078.10
|
Arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr6_+_121320339 | 2.46 |
ENSMUST00000168295.2
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr5_-_124564014 | 2.43 |
ENSMUST00000196329.5
ENSMUST00000196644.5 |
Sbno1
|
strawberry notch 1 |
| chr6_-_33037107 | 2.43 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr1_+_75119809 | 2.42 |
ENSMUST00000186037.7
ENSMUST00000187901.2 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr5_-_66308421 | 2.39 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr6_-_33037191 | 2.37 |
ENSMUST00000066379.11
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chrX_+_67722230 | 2.35 |
ENSMUST00000114656.8
|
Fmr1
|
FMRP translational regulator 1 |
| chr17_+_46694646 | 2.35 |
ENSMUST00000113481.9
ENSMUST00000138127.8 |
Zfp318
|
zinc finger protein 318 |
| chr6_+_17463748 | 2.31 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr3_-_95789338 | 2.29 |
ENSMUST00000161994.2
|
Ciart
|
circadian associated repressor of transcription |
| chr9_+_43978290 | 2.27 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr5_-_124563611 | 2.25 |
ENSMUST00000198420.5
|
Sbno1
|
strawberry notch 1 |
| chr11_-_120521382 | 2.24 |
ENSMUST00000106181.8
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr11_+_53241561 | 2.18 |
ENSMUST00000060945.12
|
Aff4
|
AF4/FMR2 family, member 4 |
| chr7_-_45109262 | 2.13 |
ENSMUST00000094434.13
|
Ftl1
|
ferritin light polypeptide 1 |
| chr2_-_48839218 | 2.08 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr3_-_95789505 | 2.07 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr16_-_4867703 | 2.06 |
ENSMUST00000115844.3
ENSMUST00000023189.15 |
Glyr1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr9_-_21913833 | 2.04 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr8_-_85573489 | 2.03 |
ENSMUST00000003912.7
|
Calr
|
calreticulin |
| chr11_+_120563844 | 2.02 |
ENSMUST00000106158.9
ENSMUST00000103016.8 ENSMUST00000168714.9 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr7_+_44499818 | 2.00 |
ENSMUST00000136232.2
ENSMUST00000207223.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr5_+_138170259 | 2.00 |
ENSMUST00000019662.11
ENSMUST00000151318.8 |
Ap4m1
|
adaptor-related protein complex AP-4, mu 1 |
| chr2_-_48839276 | 1.98 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr4_+_99544536 | 1.97 |
ENSMUST00000087285.5
|
Foxd3
|
forkhead box D3 |
| chr14_+_4230658 | 1.96 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr12_+_84498196 | 1.95 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr2_+_126549239 | 1.91 |
ENSMUST00000028841.14
ENSMUST00000110416.3 |
Usp8
|
ubiquitin specific peptidase 8 |
| chr7_-_127545896 | 1.91 |
ENSMUST00000118755.8
ENSMUST00000094026.10 |
Prss36
|
protease, serine 36 |
| chr11_+_120564185 | 1.90 |
ENSMUST00000135346.8
ENSMUST00000127269.8 ENSMUST00000131727.9 ENSMUST00000149389.8 ENSMUST00000153346.8 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr15_+_81284333 | 1.89 |
ENSMUST00000163754.9
ENSMUST00000041609.11 |
Xpnpep3
|
X-prolyl aminopeptidase 3, mitochondrial |
| chr17_+_44114894 | 1.84 |
ENSMUST00000044895.13
|
Rcan2
|
regulator of calcineurin 2 |
| chr9_-_21913896 | 1.84 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr10_-_63174801 | 1.84 |
ENSMUST00000020257.13
ENSMUST00000177694.8 ENSMUST00000105442.3 |
Sirt1
|
sirtuin 1 |
| chr13_-_64422693 | 1.83 |
ENSMUST00000109770.2
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr5_-_124563969 | 1.80 |
ENSMUST00000065263.12
|
Sbno1
|
strawberry notch 1 |
| chr2_+_155224105 | 1.77 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr6_+_143112936 | 1.76 |
ENSMUST00000204947.3
ENSMUST00000032413.7 ENSMUST00000205256.2 |
Etnk1
|
ethanolamine kinase 1 |
| chr2_-_30176324 | 1.75 |
ENSMUST00000100219.5
|
Dolk
|
dolichol kinase |
| chr2_+_91287846 | 1.74 |
ENSMUST00000028689.4
|
Lrp4
|
low density lipoprotein receptor-related protein 4 |
| chr16_-_44978546 | 1.74 |
ENSMUST00000114600.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr3_+_96542746 | 1.71 |
ENSMUST00000118557.8
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr5_-_123822351 | 1.68 |
ENSMUST00000111564.8
ENSMUST00000063905.12 |
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr3_+_96543143 | 1.68 |
ENSMUST00000165842.3
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr3_+_96542933 | 1.65 |
ENSMUST00000048766.15
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr1_+_191638854 | 1.65 |
ENSMUST00000044954.7
|
Slc30a1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr2_+_48839505 | 1.65 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr5_-_113285852 | 1.65 |
ENSMUST00000212276.2
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chrX_+_165127688 | 1.64 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr5_-_124563636 | 1.64 |
ENSMUST00000196711.5
ENSMUST00000200474.5 ENSMUST00000199808.5 |
Sbno1
|
strawberry notch 1 |
| chr2_+_155118217 | 1.63 |
ENSMUST00000029128.4
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr1_-_10302895 | 1.62 |
ENSMUST00000088615.11
ENSMUST00000131556.2 |
Arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) |
| chr19_-_60456742 | 1.61 |
ENSMUST00000051277.4
|
Prlhr
|
prolactin releasing hormone receptor |
| chr6_+_17463819 | 1.61 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr9_+_43978369 | 1.60 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr16_-_5021843 | 1.58 |
ENSMUST00000147567.2
ENSMUST00000023911.11 |
Nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr2_+_19662529 | 1.58 |
ENSMUST00000052168.6
|
Otud1
|
OTU domain containing 1 |
| chr4_-_129083392 | 1.57 |
ENSMUST00000117497.8
ENSMUST00000117350.2 |
S100pbp
|
S100P binding protein |
| chr11_+_117545037 | 1.57 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr15_-_53209513 | 1.56 |
ENSMUST00000077273.9
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr1_+_171173252 | 1.56 |
ENSMUST00000006579.5
|
Pfdn2
|
prefoldin 2 |
| chr4_-_129083439 | 1.55 |
ENSMUST00000106059.8
|
S100pbp
|
S100P binding protein |
| chr2_-_37593287 | 1.51 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr5_+_24633206 | 1.48 |
ENSMUST00000115049.9
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr7_-_129867967 | 1.48 |
ENSMUST00000117872.8
ENSMUST00000120187.9 |
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr16_-_44978816 | 1.48 |
ENSMUST00000181750.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chrX_+_70860378 | 1.47 |
ENSMUST00000114575.4
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr14_+_30741115 | 1.47 |
ENSMUST00000112094.8
ENSMUST00000144009.2 |
Pbrm1
|
polybromo 1 |
| chr5_-_123822338 | 1.46 |
ENSMUST00000111561.8
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr11_-_101062111 | 1.46 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr1_+_131935342 | 1.45 |
ENSMUST00000086556.12
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr2_+_3514071 | 1.39 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_-_183078800 | 1.39 |
ENSMUST00000163528.8
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr7_+_44499374 | 1.38 |
ENSMUST00000141311.8
ENSMUST00000107880.9 ENSMUST00000208384.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr1_-_155617773 | 1.38 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr19_-_42117420 | 1.37 |
ENSMUST00000161873.2
ENSMUST00000018965.4 |
Avpi1
|
arginine vasopressin-induced 1 |
| chr17_-_65920481 | 1.36 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr10_+_126899396 | 1.36 |
ENSMUST00000006911.12
|
Cdk4
|
cyclin-dependent kinase 4 |
| chr14_-_72840373 | 1.35 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr1_-_20854490 | 1.35 |
ENSMUST00000039046.10
|
Il17f
|
interleukin 17F |
| chr7_+_27878894 | 1.35 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr7_+_125307060 | 1.35 |
ENSMUST00000124223.8
ENSMUST00000069660.13 |
Katnip
|
katanin interacting protein |
| chr11_+_117545618 | 1.34 |
ENSMUST00000106344.8
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr10_-_17823736 | 1.31 |
ENSMUST00000037879.8
|
Heca
|
hdc homolog, cell cycle regulator |
| chr4_+_48585275 | 1.29 |
ENSMUST00000123476.8
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr1_-_130643471 | 1.28 |
ENSMUST00000066863.13
ENSMUST00000169659.8 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr13_-_64300964 | 1.27 |
ENSMUST00000059817.12
ENSMUST00000117241.2 |
Zfp367
|
zinc finger protein 367 |
| chrX_+_70860357 | 1.26 |
ENSMUST00000114576.9
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr11_-_101061153 | 1.25 |
ENSMUST00000123864.2
|
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr7_+_121666388 | 1.25 |
ENSMUST00000033158.6
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr6_+_7555053 | 1.24 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr1_+_87254729 | 1.23 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr1_-_30912662 | 1.19 |
ENSMUST00000186733.7
|
Phf3
|
PHD finger protein 3 |
| chr10_-_128759331 | 1.17 |
ENSMUST00000153731.8
ENSMUST00000026405.10 |
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr10_+_121200984 | 1.15 |
ENSMUST00000040344.7
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr8_+_3550533 | 1.15 |
ENSMUST00000208306.2
|
Mcoln1
|
mucolipin 1 |
| chr2_+_160573604 | 1.15 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr8_+_3550450 | 1.14 |
ENSMUST00000004683.13
ENSMUST00000160338.2 |
Mcoln1
|
mucolipin 1 |
| chr10_+_98750268 | 1.14 |
ENSMUST00000219557.2
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr9_-_91247831 | 1.13 |
ENSMUST00000065360.5
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr7_-_121666486 | 1.11 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr14_+_30741082 | 1.11 |
ENSMUST00000112098.11
ENSMUST00000112095.8 ENSMUST00000112106.8 ENSMUST00000146325.8 |
Pbrm1
|
polybromo 1 |
| chr3_-_89820451 | 1.11 |
ENSMUST00000029559.7
ENSMUST00000197679.5 |
Il6ra
|
interleukin 6 receptor, alpha |
| chr1_-_30912916 | 1.09 |
ENSMUST00000188780.2
|
Phf3
|
PHD finger protein 3 |
| chr7_-_46445085 | 1.07 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr13_-_92268156 | 1.07 |
ENSMUST00000151408.9
ENSMUST00000216219.2 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr9_-_91247809 | 1.07 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr12_+_4284009 | 1.04 |
ENSMUST00000179139.3
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr4_-_126096112 | 1.03 |
ENSMUST00000142125.2
ENSMUST00000106141.3 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chrX_+_67722147 | 1.03 |
ENSMUST00000088546.12
|
Fmr1
|
FMRP translational regulator 1 |
| chr7_-_46445305 | 1.02 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr17_-_56916771 | 1.02 |
ENSMUST00000052832.6
|
Micos13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr4_-_119279551 | 1.01 |
ENSMUST00000106316.2
ENSMUST00000030385.13 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr6_+_116184991 | 1.01 |
ENSMUST00000036759.11
ENSMUST00000204476.3 |
Washc2
|
WASH complex subunit 2` |
| chr5_-_66308666 | 0.99 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr9_+_59446823 | 0.99 |
ENSMUST00000026262.8
|
Hexa
|
hexosaminidase A |
| chr6_-_108162513 | 0.97 |
ENSMUST00000167338.8
ENSMUST00000172188.2 ENSMUST00000032191.16 |
Sumf1
|
sulfatase modifying factor 1 |
| chr12_-_85421467 | 0.97 |
ENSMUST00000040766.9
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr1_+_183078573 | 0.97 |
ENSMUST00000109166.8
|
Aida
|
axin interactor, dorsalization associated |
| chr10_-_127504416 | 0.97 |
ENSMUST00000129252.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr1_-_43131598 | 0.94 |
ENSMUST00000188728.2
|
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr4_-_129083335 | 0.93 |
ENSMUST00000106061.9
ENSMUST00000072431.13 |
S100pbp
|
S100P binding protein |
| chr14_+_30741140 | 0.92 |
ENSMUST00000052239.12
|
Pbrm1
|
polybromo 1 |
| chr2_+_180570793 | 0.92 |
ENSMUST00000108875.2
|
Birc7
|
baculoviral IAP repeat-containing 7 (livin) |
| chr7_+_16043502 | 0.91 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr11_-_83320281 | 0.91 |
ENSMUST00000052521.9
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr15_+_59520199 | 0.91 |
ENSMUST00000067543.8
|
Trib1
|
tribbles pseudokinase 1 |
| chr15_+_59520493 | 0.89 |
ENSMUST00000118228.2
|
Trib1
|
tribbles pseudokinase 1 |
| chr15_-_93234681 | 0.87 |
ENSMUST00000080299.7
|
Yaf2
|
YY1 associated factor 2 |
| chr16_+_17577464 | 0.86 |
ENSMUST00000129199.8
|
Klhl22
|
kelch-like 22 |
| chr5_-_24745970 | 0.86 |
ENSMUST00000117900.8
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr17_-_67939702 | 0.85 |
ENSMUST00000097290.4
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr11_-_118931731 | 0.84 |
ENSMUST00000026663.8
|
Cbx8
|
chromobox 8 |
| chr5_-_65855199 | 0.84 |
ENSMUST00000031104.7
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr2_+_74557418 | 0.84 |
ENSMUST00000111980.4
|
Hoxd4
|
homeobox D4 |
| chr17_+_34043536 | 0.84 |
ENSMUST00000048249.8
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr7_+_125307116 | 0.83 |
ENSMUST00000148701.4
|
Katnip
|
katanin interacting protein |
| chr4_+_126971167 | 0.81 |
ENSMUST00000046751.13
ENSMUST00000094713.4 |
Zmym6
|
zinc finger, MYM-type 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 2.0 | 10.0 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.9 | 5.8 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.4 | 5.4 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 1.3 | 3.9 | GO:1901254 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 1.1 | 7.7 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.0 | 5.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.0 | 3.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.9 | 2.8 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.9 | 6.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.9 | 3.5 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.9 | 2.6 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.8 | 2.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.7 | 2.2 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 0.7 | 3.5 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.7 | 2.0 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.6 | 3.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.6 | 1.8 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.6 | 2.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.6 | 1.7 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.5 | 3.0 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.5 | 2.5 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.5 | 3.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 1.4 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.5 | 0.5 | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) |
| 0.5 | 5.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 3.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.4 | 1.7 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.4 | 2.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 2.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.4 | 4.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.4 | 1.2 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.4 | 1.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 | 6.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 1.6 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.4 | 2.8 | GO:0072719 | cellular response to cisplatin(GO:0072719) copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.4 | 10.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.4 | 1.4 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.3 | 4.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 1.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.3 | 1.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 1.4 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.2 | 3.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 8.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 4.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 5.5 | GO:0044804 | nucleophagy(GO:0044804) negative regulation of phagocytosis(GO:0050765) |
| 0.2 | 1.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 2.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.0 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.2 | 0.9 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 2.9 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 3.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 1.4 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 5.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.2 | 6.0 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.2 | 0.8 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.2 | 1.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.2 | 0.6 | GO:0061055 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.2 | 1.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.6 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 2.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.4 | GO:0060112 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 0.6 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 1.8 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 1.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.5 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 3.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 2.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 1.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.3 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.1 | 1.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 1.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.6 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 2.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 1.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 7.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 0.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.5 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 17.0 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 1.8 | GO:0007614 | short-term memory(GO:0007614) locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 2.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 5.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 2.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.1 | GO:0070050 | negative thymic T cell selection(GO:0045060) neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 1.8 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.5 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 1.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.9 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 2.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.9 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 4.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 2.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.4 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.9 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 2.2 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.8 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 1.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.7 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 1.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 2.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.6 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.7 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 1.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 2.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0060394 | maintenance of gastrointestinal epithelium(GO:0030277) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 1.1 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.7 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 1.9 | 5.8 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.0 | 3.9 | GO:0019034 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.6 | 25.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.6 | 2.5 | GO:0005879 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.6 | 4.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 2.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.4 | 4.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 5.8 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.3 | 3.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 3.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 1.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 3.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 1.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 2.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 3.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 1.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 2.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.2 | 3.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 4.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 5.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.9 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 2.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.6 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 3.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 3.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 7.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 13.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 20.5 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 3.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 4.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 4.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 4.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.9 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.8 | 5.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 1.7 | 10.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.7 | 6.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.7 | 2.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.6 | 3.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.6 | 1.8 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.6 | 5.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 1.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.5 | 3.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.5 | 3.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.5 | 12.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.5 | 6.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 2.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 1.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.4 | 1.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.4 | 3.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.8 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.4 | 2.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 0.8 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.2 | 5.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 4.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 1.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 4.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 2.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 1.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 4.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 0.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 2.8 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 10.9 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.2 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 3.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 2.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 3.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 3.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.5 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 2.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.6 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 1.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 4.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 7.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 8.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.2 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 4.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 2.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 2.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 4.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 3.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 1.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 3.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 2.1 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 7.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 4.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 4.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 3.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 4.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.3 | 5.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 3.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 4.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 3.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 8.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 3.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 2.2 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.2 | 2.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 4.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 4.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 4.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 4.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 2.9 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 2.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.8 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 5.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 4.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |