Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
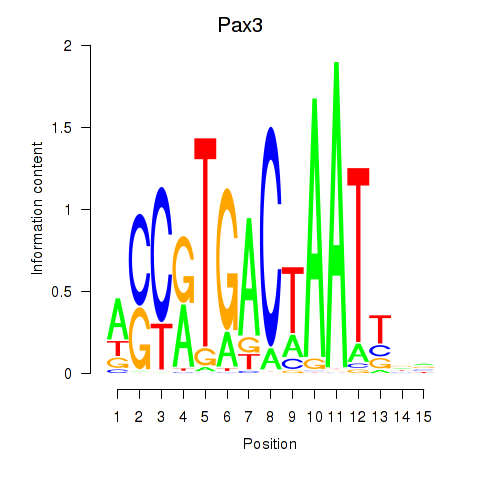
Results for Pax3
Z-value: 1.55
Transcription factors associated with Pax3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax3
|
ENSMUSG00000004872.16 | Pax3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax3 | mm39_v1_chr1_-_78173468_78173482 | 0.01 | 9.4e-01 | Click! |
Activity profile of Pax3 motif
Sorted Z-values of Pax3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_84058292 | 10.90 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr5_-_87240405 | 10.61 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr13_+_4484305 | 10.55 |
ENSMUST00000021630.15
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr2_+_102489558 | 6.41 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_-_46136765 | 2.87 |
ENSMUST00000026259.16
|
Pitx3
|
paired-like homeodomain transcription factor 3 |
| chr10_-_115197775 | 2.87 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr7_-_133384449 | 2.28 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr19_-_36714053 | 2.12 |
ENSMUST00000087321.4
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr11_-_88742285 | 2.02 |
ENSMUST00000107903.8
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr10_-_128759331 | 1.64 |
ENSMUST00000153731.8
ENSMUST00000026405.10 |
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr10_-_128759817 | 1.63 |
ENSMUST00000131271.2
|
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr1_+_176642226 | 1.41 |
ENSMUST00000056773.15
ENSMUST00000027785.15 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr4_-_25281750 | 1.31 |
ENSMUST00000038705.8
ENSMUST00000102994.10 |
Ufl1
|
UFM1 specific ligase 1 |
| chr3_+_108093645 | 1.07 |
ENSMUST00000050909.7
ENSMUST00000106659.3 ENSMUST00000106656.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr16_+_20367327 | 0.98 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr4_+_126971167 | 0.85 |
ENSMUST00000046751.13
ENSMUST00000094713.4 |
Zmym6
|
zinc finger, MYM-type 6 |
| chr8_+_111345209 | 0.84 |
ENSMUST00000034190.11
|
Vac14
|
Vac14 homolog (S. cerevisiae) |
| chr10_-_80374916 | 0.83 |
ENSMUST00000219648.2
|
Atp8b3
|
ATPase, class I, type 8B, member 3 |
| chr14_+_52131067 | 0.83 |
ENSMUST00000047726.12
|
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr2_+_133394079 | 0.79 |
ENSMUST00000028836.7
|
Bmp2
|
bone morphogenetic protein 2 |
| chr3_-_59170245 | 0.66 |
ENSMUST00000050360.14
ENSMUST00000199609.2 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr4_+_106954794 | 0.65 |
ENSMUST00000221740.2
|
Cdcp2
|
CUB domain containing protein 2 |
| chr6_+_34897904 | 0.61 |
ENSMUST00000185102.2
ENSMUST00000114997.3 |
Stra8
|
stimulated by retinoic acid gene 8 |
| chr13_-_21900313 | 0.58 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr13_-_74538899 | 0.54 |
ENSMUST00000223163.3
|
Zfp72
|
zinc finger protein 72 |
| chr2_+_168072519 | 0.54 |
ENSMUST00000099071.5
|
Mocs3
|
molybdenum cofactor synthesis 3 |
| chr10_+_36382810 | 0.52 |
ENSMUST00000167191.8
ENSMUST00000058738.11 |
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr16_+_4964849 | 0.49 |
ENSMUST00000165810.2
ENSMUST00000230616.2 |
Sec14l5
|
SEC14-like lipid binding 5 |
| chr12_-_72231802 | 0.45 |
ENSMUST00000021494.6
|
Ccdc175
|
coiled-coil domain containing 175 |
| chr7_-_139941566 | 0.43 |
ENSMUST00000215023.2
ENSMUST00000216027.2 ENSMUST00000210932.3 ENSMUST00000211031.3 |
Olfr60
|
olfactory receptor 60 |
| chr2_-_136229849 | 0.41 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr17_-_23990512 | 0.40 |
ENSMUST00000226460.2
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chrX_+_113384008 | 0.40 |
ENSMUST00000113371.8
ENSMUST00000040504.12 |
Klhl4
|
kelch-like 4 |
| chr16_-_32698091 | 0.38 |
ENSMUST00000119810.2
|
Rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr10_+_103987773 | 0.37 |
ENSMUST00000180692.3
|
Gm6763
|
predicted gene 6763 |
| chr10_+_103996282 | 0.37 |
ENSMUST00000181166.3
|
Gm8764
|
predicted gene 8764 |
| chr10_+_104030318 | 0.37 |
ENSMUST00000181036.2
|
Gm4340
|
predicted gene 4340 |
| chr10_+_103979263 | 0.37 |
ENSMUST00000180664.2
|
Gm21293
|
predicted gene, 21293 |
| chr10_+_104004791 | 0.37 |
ENSMUST00000180568.2
|
Gm21304
|
predicted gene, 21304 |
| chr10_+_104013300 | 0.37 |
ENSMUST00000181239.2
|
Gm21312
|
predicted gene, 21312 |
| chr10_+_104021809 | 0.37 |
ENSMUST00000181707.2
|
Gm20765
|
predicted gene, 20765 |
| chr6_-_87312681 | 0.36 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr11_+_3599183 | 0.34 |
ENSMUST00000096441.5
|
Morc2a
|
microrchidia 2A |
| chr3_-_75359125 | 0.31 |
ENSMUST00000204341.3
|
Wdr49
|
WD repeat domain 49 |
| chr3_+_79791798 | 0.29 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr19_+_33985684 | 0.29 |
ENSMUST00000225505.2
|
Lipk
|
lipase, family member K |
| chr6_+_34897874 | 0.28 |
ENSMUST00000114999.8
|
Stra8
|
stimulated by retinoic acid gene 8 |
| chr1_-_130815144 | 0.28 |
ENSMUST00000121040.8
|
Il24
|
interleukin 24 |
| chr19_+_33985701 | 0.28 |
ENSMUST00000054260.8
|
Lipk
|
lipase, family member K |
| chr11_-_3599673 | 0.28 |
ENSMUST00000193809.2
|
Tug1
|
taurine upregulated gene 1 |
| chr10_-_70428611 | 0.27 |
ENSMUST00000162251.8
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr17_-_23990479 | 0.27 |
ENSMUST00000086325.13
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr10_-_107747995 | 0.27 |
ENSMUST00000165341.5
|
Otogl
|
otogelin-like |
| chr3_-_33136153 | 0.26 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr9_-_40015750 | 0.26 |
ENSMUST00000213858.2
|
Olfr984
|
olfactory receptor 984 |
| chr4_+_109092829 | 0.24 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chr4_-_63663344 | 0.23 |
ENSMUST00000239480.2
ENSMUST00000062246.8 |
Tnfsf15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chrX_+_156485570 | 0.22 |
ENSMUST00000112520.2
|
Smpx
|
small muscle protein, X-linked |
| chr10_+_104029903 | 0.21 |
ENSMUST00000181615.8
|
Gm4340
|
predicted gene 4340 |
| chr10_+_103978848 | 0.21 |
ENSMUST00000181634.8
|
Gm21293
|
predicted gene, 21293 |
| chr10_+_103987358 | 0.21 |
ENSMUST00000181287.5
|
Gm6763
|
predicted gene 6763 |
| chr10_+_103995867 | 0.21 |
ENSMUST00000181179.5
|
Gm8764
|
predicted gene 8764 |
| chr10_+_104004376 | 0.21 |
ENSMUST00000181059.8
|
Gm21304
|
predicted gene, 21304 |
| chr10_+_104012885 | 0.21 |
ENSMUST00000180889.8
|
Gm21312
|
predicted gene, 21312 |
| chr10_+_104021394 | 0.21 |
ENSMUST00000181703.8
|
Gm20765
|
predicted gene, 20765 |
| chr12_+_38830812 | 0.20 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr4_+_115099237 | 0.17 |
ENSMUST00000118278.2
|
Cyp4a29
|
cytochrome P450, family 4, subfamily a, polypeptide 29 |
| chr6_+_43083303 | 0.16 |
ENSMUST00000213649.2
|
Olfr441
|
olfactory receptor 441 |
| chr13_+_14804739 | 0.14 |
ENSMUST00000178289.3
ENSMUST00000038690.6 ENSMUST00000222052.2 |
AW209491
|
expressed sequence AW209491 |
| chr11_-_52165682 | 0.13 |
ENSMUST00000238914.2
|
Tcf7
|
transcription factor 7, T cell specific |
| chr12_+_38830283 | 0.11 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chr18_+_35695199 | 0.10 |
ENSMUST00000236860.2
ENSMUST00000166793.10 ENSMUST00000237780.2 ENSMUST00000236507.2 ENSMUST00000235960.2 ENSMUST00000237061.2 |
Matr3
|
matrin 3 |
| chr12_+_38831093 | 0.07 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr9_-_35559489 | 0.03 |
ENSMUST00000178236.3
|
Pate3
|
prostate and testis expressed 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.0 | 2.9 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.9 | 6.4 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.3 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 2.0 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.8 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.2 | 0.7 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 3.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.9 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.5 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 1.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 2.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 3.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 6.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.8 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.1 | 6.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 10.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.5 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 2.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 6.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |