Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pax4
Z-value: 1.27
Transcription factors associated with Pax4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax4
|
ENSMUSG00000029706.16 | Pax4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax4 | mm39_v1_chr6_-_28447179_28447179 | 0.26 | 1.3e-01 | Click! |
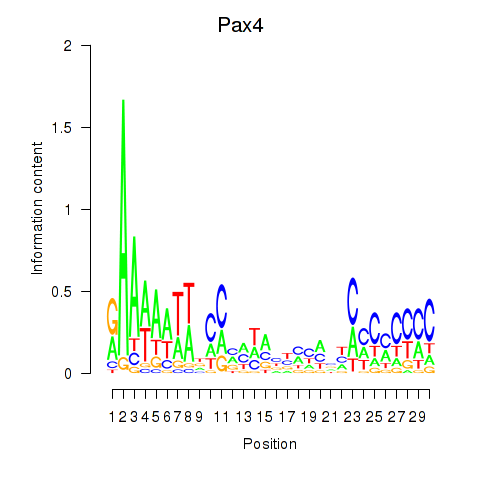
Activity profile of Pax4 motif
Sorted Z-values of Pax4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_59563872 | 5.31 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr9_+_59564482 | 5.14 |
ENSMUST00000216620.2
ENSMUST00000217038.2 |
Pkm
|
pyruvate kinase, muscle |
| chr9_+_59563838 | 5.03 |
ENSMUST00000163694.4
|
Pkm
|
pyruvate kinase, muscle |
| chr7_+_78563150 | 4.39 |
ENSMUST00000118867.8
|
Isg20
|
interferon-stimulated protein |
| chr7_+_78563513 | 4.35 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr7_+_78563184 | 4.11 |
ENSMUST00000121645.8
|
Isg20
|
interferon-stimulated protein |
| chr11_+_98798627 | 4.00 |
ENSMUST00000092706.13
|
Cdc6
|
cell division cycle 6 |
| chr17_+_35278011 | 3.91 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_62465304 | 3.30 |
ENSMUST00000018651.14
|
Trpv2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr16_-_36486429 | 3.30 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr7_+_24561616 | 2.63 |
ENSMUST00000170837.3
|
Gm9844
|
predicted pseudogene 9844 |
| chr13_-_55676334 | 2.57 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr5_+_66833434 | 2.21 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr11_+_106916430 | 2.07 |
ENSMUST00000140447.4
|
1810010H24Rik
|
RIKEN cDNA 1810010H24 gene |
| chr9_+_32607301 | 2.05 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr5_+_135703426 | 2.00 |
ENSMUST00000153515.8
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr17_-_23864237 | 1.94 |
ENSMUST00000024696.9
|
Mmp25
|
matrix metallopeptidase 25 |
| chr2_-_125348305 | 1.86 |
ENSMUST00000028633.13
|
Fbn1
|
fibrillin 1 |
| chr4_-_87951565 | 1.84 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr15_+_78481247 | 1.83 |
ENSMUST00000043069.6
ENSMUST00000231180.2 ENSMUST00000229796.2 ENSMUST00000229295.2 |
Cyth4
|
cytohesin 4 |
| chr9_+_96141317 | 1.80 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr9_+_96141299 | 1.77 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr4_-_58987094 | 1.64 |
ENSMUST00000030069.7
|
Ptgr1
|
prostaglandin reductase 1 |
| chr1_+_153628598 | 1.61 |
ENSMUST00000182538.3
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr6_+_47854138 | 1.60 |
ENSMUST00000061890.8
|
Zfp282
|
zinc finger protein 282 |
| chr4_+_6365694 | 1.54 |
ENSMUST00000175769.8
ENSMUST00000140830.8 ENSMUST00000108374.8 |
Sdcbp
|
syndecan binding protein |
| chr19_-_4240984 | 1.50 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr16_-_14109219 | 1.46 |
ENSMUST00000230397.2
ENSMUST00000231567.2 ENSMUST00000090287.5 |
Myh11
|
myosin, heavy polypeptide 11, smooth muscle |
| chr19_+_53128861 | 1.40 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr9_-_58066484 | 1.34 |
ENSMUST00000041477.15
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr4_+_6365650 | 1.30 |
ENSMUST00000029912.11
ENSMUST00000103008.12 |
Sdcbp
|
syndecan binding protein |
| chr12_-_84265609 | 1.24 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr2_+_22785534 | 1.20 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr1_-_79838897 | 1.18 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr7_+_43057611 | 1.17 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr2_+_22785593 | 0.96 |
ENSMUST00000152170.8
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr19_-_4241034 | 0.92 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr8_-_83129134 | 0.87 |
ENSMUST00000209363.2
|
Il15
|
interleukin 15 |
| chr4_+_150366028 | 0.87 |
ENSMUST00000105682.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr13_+_23922783 | 0.82 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr2_-_160155536 | 0.77 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr4_+_124960465 | 0.76 |
ENSMUST00000052183.7
|
Snip1
|
Smad nuclear interacting protein 1 |
| chr3_+_95532282 | 0.74 |
ENSMUST00000058230.13
ENSMUST00000037983.6 |
Ensa
|
endosulfine alpha |
| chr7_+_80764564 | 0.69 |
ENSMUST00000119083.2
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chrX_+_72235828 | 0.69 |
ENSMUST00000101486.5
|
Xlr3b
|
X-linked lymphocyte-regulated 3B |
| chr13_+_21901791 | 0.66 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr11_+_87017878 | 0.66 |
ENSMUST00000041282.13
|
Trim37
|
tripartite motif-containing 37 |
| chr11_-_18968955 | 0.66 |
ENSMUST00000068264.14
ENSMUST00000185131.8 |
Meis1
|
Meis homeobox 1 |
| chr19_+_4022322 | 0.63 |
ENSMUST00000143380.3
|
Aldh3b2
|
aldehyde dehydrogenase 3 family, member B2 |
| chr2_-_114485421 | 0.62 |
ENSMUST00000028640.14
ENSMUST00000102542.10 |
Dph6
|
diphthamine biosynthesis 6 |
| chr7_+_80764547 | 0.61 |
ENSMUST00000026820.11
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr4_+_147216495 | 0.54 |
ENSMUST00000084149.10
|
Zfp991
|
zinc finger protein 991 |
| chr8_-_83128992 | 0.53 |
ENSMUST00000210094.2
|
Il15
|
interleukin 15 |
| chr10_+_119655294 | 0.53 |
ENSMUST00000105262.9
ENSMUST00000147454.8 ENSMUST00000138410.8 ENSMUST00000144825.8 ENSMUST00000148954.8 ENSMUST00000144959.8 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_+_181322077 | 0.52 |
ENSMUST00000103042.10
|
Tcea2
|
transcription elongation factor A (SII), 2 |
| chr6_+_136495784 | 0.49 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr13_-_101904662 | 0.43 |
ENSMUST00000055518.13
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr10_+_129153986 | 0.43 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr2_-_114485342 | 0.43 |
ENSMUST00000055144.8
|
Dph6
|
diphthamine biosynthesis 6 |
| chr15_-_59245998 | 0.42 |
ENSMUST00000022976.6
|
Washc5
|
WASH complex subunit 5 |
| chr10_+_127257077 | 0.37 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr2_-_131170902 | 0.35 |
ENSMUST00000110194.8
|
Rnf24
|
ring finger protein 24 |
| chr9_+_14411907 | 0.34 |
ENSMUST00000004200.9
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr5_-_121641461 | 0.33 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr12_+_71095112 | 0.32 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr10_-_129509659 | 0.32 |
ENSMUST00000213294.2
ENSMUST00000216067.3 ENSMUST00000203424.2 |
Olfr801
|
olfactory receptor 801 |
| chr9_+_88209250 | 0.31 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr9_+_20209828 | 0.29 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr10_+_89906956 | 0.28 |
ENSMUST00000183109.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_+_136011969 | 0.27 |
ENSMUST00000144217.8
|
Zfp46
|
zinc finger protein 46 |
| chr17_+_69690314 | 0.26 |
ENSMUST00000062369.14
|
Zbtb14
|
zinc finger and BTB domain containing 14 |
| chr5_+_124690908 | 0.26 |
ENSMUST00000071057.14
ENSMUST00000111438.2 |
Ddx55
|
DEAD box helicase 55 |
| chr10_+_127256993 | 0.25 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chrM_+_11735 | 0.23 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr19_+_55169148 | 0.22 |
ENSMUST00000154886.8
ENSMUST00000120936.8 ENSMUST00000025936.12 |
Tectb
|
tectorin beta |
| chr17_+_47604995 | 0.21 |
ENSMUST00000190020.4
|
Trerf1
|
transcriptional regulating factor 1 |
| chr6_+_136495818 | 0.21 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_+_43090206 | 0.20 |
ENSMUST00000040227.3
|
Cldnd2
|
claudin domain containing 2 |
| chr10_+_21870565 | 0.15 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr9_-_101076198 | 0.14 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr9_+_36743980 | 0.09 |
ENSMUST00000034630.15
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chrX_-_153911405 | 0.09 |
ENSMUST00000076671.4
|
Cldn34b2
|
claudin 34B2 |
| chr17_+_69690018 | 0.08 |
ENSMUST00000112674.8
|
Zbtb14
|
zinc finger and BTB domain containing 14 |
| chr4_+_146695418 | 0.05 |
ENSMUST00000130825.8
|
Zfp993
|
zinc finger protein 993 |
| chr17_+_17622934 | 0.03 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr3_+_99147677 | 0.03 |
ENSMUST00000151606.8
|
Tbx15
|
T-box 15 |
| chr17_+_34457868 | 0.01 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr16_-_13548307 | 0.01 |
ENSMUST00000115807.9
|
Pla2g10
|
phospholipase A2, group X |
| chrX_-_52298601 | 0.01 |
ENSMUST00000174390.2
|
Plac1
|
placental specific protein 1 |
| chr16_-_45664591 | 0.00 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.5 | 15.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.5 | 2.0 | GO:0090345 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.5 | 2.8 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.5 | 1.9 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.4 | 1.2 | GO:0042628 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) seminal vesicle development(GO:0061107) |
| 0.4 | 2.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 1.3 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.3 | 3.9 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.3 | 2.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 1.9 | GO:0060022 | hard palate development(GO:0060022) |
| 0.2 | 1.4 | GO:0045062 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.2 | 3.3 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.2 | 0.9 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 2.1 | GO:0030578 | PML body organization(GO:0030578) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 4.0 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.1 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 2.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.3 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.1 | 1.6 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 0.4 | GO:0090306 | polar body extrusion after meiotic divisions(GO:0040038) spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 1.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 3.3 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.6 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.8 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.3 | 3.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 12.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 4.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 2.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 1.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 3.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.6 | 12.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.9 | 2.8 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.6 | 2.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 2.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.5 | 2.0 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.4 | 1.3 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.4 | 1.6 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.4 | 3.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.6 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 1.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 2.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 3.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 3.6 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 14.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 3.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |