Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
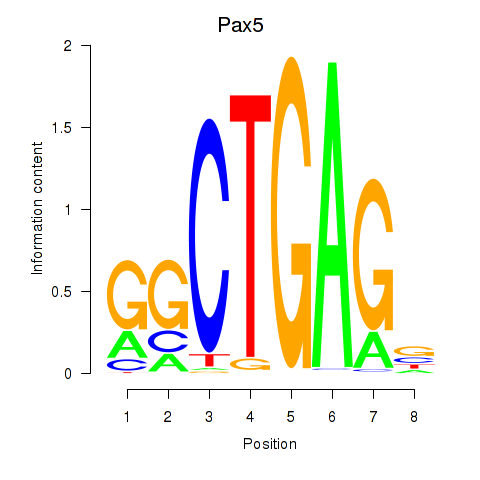
Results for Pax5
Z-value: 1.57
Transcription factors associated with Pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax5
|
ENSMUSG00000014030.16 | Pax5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax5 | mm39_v1_chr4_-_44645723_44645723 | 0.11 | 5.4e-01 | Click! |
Activity profile of Pax5 motif
Sorted Z-values of Pax5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142233270 | 6.63 |
ENSMUST00000162317.2
ENSMUST00000125933.2 ENSMUST00000105931.8 ENSMUST00000105930.8 ENSMUST00000105933.8 ENSMUST00000105932.2 ENSMUST00000000220.3 |
Ins2
|
insulin II |
| chr9_+_46180362 | 5.31 |
ENSMUST00000214202.2
ENSMUST00000215458.2 ENSMUST00000215187.2 ENSMUST00000213878.2 ENSMUST00000034584.4 |
Apoa5
|
apolipoprotein A-V |
| chr19_+_52252735 | 4.76 |
ENSMUST00000039652.6
|
Ins1
|
insulin I |
| chr6_+_30541581 | 4.53 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr10_+_127612243 | 4.32 |
ENSMUST00000136223.2
ENSMUST00000052652.7 |
Rdh9
|
retinol dehydrogenase 9 |
| chr10_+_127595639 | 3.72 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr11_+_69945157 | 3.21 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr19_+_3373285 | 3.01 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr18_+_9212154 | 2.95 |
ENSMUST00000041080.7
|
Fzd8
|
frizzled class receptor 8 |
| chr15_+_76580925 | 2.87 |
ENSMUST00000023203.6
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr10_-_128796834 | 2.81 |
ENSMUST00000026398.5
|
Mettl7b
|
methyltransferase like 7B |
| chr4_+_41569775 | 2.35 |
ENSMUST00000102963.10
|
Dnai1
|
dynein axonemal intermediate chain 1 |
| chr5_-_134776101 | 2.32 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr16_+_22710134 | 2.25 |
ENSMUST00000231328.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr16_+_22710785 | 2.14 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr10_-_127724557 | 2.11 |
ENSMUST00000047199.5
|
Rdh7
|
retinol dehydrogenase 7 |
| chr16_+_22710027 | 2.10 |
ENSMUST00000231848.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr10_+_127702326 | 2.09 |
ENSMUST00000092058.4
|
Rdh16f2
|
RDH16 family member 2 |
| chr4_-_107164347 | 2.05 |
ENSMUST00000082426.11
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr4_+_115420876 | 2.04 |
ENSMUST00000126645.8
ENSMUST00000030480.4 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr4_-_107164315 | 2.02 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr7_+_130633776 | 1.97 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr13_+_58956077 | 1.94 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr10_-_7831657 | 1.92 |
ENSMUST00000147938.2
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr16_+_44914397 | 1.90 |
ENSMUST00000061050.6
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr16_+_44913974 | 1.89 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr8_+_105652867 | 1.82 |
ENSMUST00000034355.11
ENSMUST00000109410.4 |
Ces2e
|
carboxylesterase 2E |
| chr6_+_134807097 | 1.77 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr10_+_127637015 | 1.74 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr5_+_89175815 | 1.73 |
ENSMUST00000130041.8
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr13_+_58956495 | 1.72 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr14_+_55813074 | 1.61 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr11_+_98938137 | 1.59 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr11_-_101998648 | 1.59 |
ENSMUST00000177304.8
ENSMUST00000017455.15 |
Pyy
|
peptide YY |
| chr6_+_34723304 | 1.55 |
ENSMUST00000142716.3
|
Cald1
|
caldesmon 1 |
| chr18_-_60866186 | 1.54 |
ENSMUST00000237885.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr8_-_94738748 | 1.52 |
ENSMUST00000143265.2
|
Amfr
|
autocrine motility factor receptor |
| chr19_-_58442866 | 1.51 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr1_+_88093726 | 1.50 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr2_+_165834546 | 1.49 |
ENSMUST00000109252.8
ENSMUST00000088095.6 |
Ncoa3
|
nuclear receptor coactivator 3 |
| chrX_-_59449137 | 1.49 |
ENSMUST00000033480.13
ENSMUST00000101527.3 |
Atp11c
|
ATPase, class VI, type 11C |
| chr8_+_46944000 | 1.48 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr14_+_21102662 | 1.48 |
ENSMUST00000223915.2
|
Adk
|
adenosine kinase |
| chr2_-_104324035 | 1.47 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr7_-_12731594 | 1.46 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr7_-_12732067 | 1.41 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr12_-_91815855 | 1.40 |
ENSMUST00000167466.2
ENSMUST00000021347.12 ENSMUST00000178462.8 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr19_-_58443012 | 1.40 |
ENSMUST00000129100.8
ENSMUST00000123957.2 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr14_-_30645503 | 1.39 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr6_+_34722887 | 1.36 |
ENSMUST00000123823.8
ENSMUST00000136907.8 |
Cald1
|
caldesmon 1 |
| chr3_-_116762617 | 1.36 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr13_-_55574596 | 1.36 |
ENSMUST00000021948.15
|
F12
|
coagulation factor XII (Hageman factor) |
| chr19_+_25384024 | 1.36 |
ENSMUST00000146647.3
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr16_-_46317318 | 1.35 |
ENSMUST00000023335.13
ENSMUST00000023334.15 |
Nectin3
|
nectin cell adhesion molecule 3 |
| chr2_-_32341408 | 1.35 |
ENSMUST00000028160.15
ENSMUST00000113310.9 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr4_-_149391963 | 1.33 |
ENSMUST00000055647.15
ENSMUST00000030806.6 ENSMUST00000238956.2 ENSMUST00000060537.13 |
Kif1b
|
kinesin family member 1B |
| chr19_-_7780025 | 1.31 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr6_+_90310252 | 1.29 |
ENSMUST00000046128.12
ENSMUST00000164761.6 |
Uroc1
|
urocanase domain containing 1 |
| chr7_+_140500848 | 1.28 |
ENSMUST00000184560.2
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr11_-_116080361 | 1.27 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr5_-_122639840 | 1.26 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr9_-_107544573 | 1.25 |
ENSMUST00000010208.14
ENSMUST00000193932.6 |
Slc38a3
|
solute carrier family 38, member 3 |
| chr3_+_94284739 | 1.24 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr4_-_41503046 | 1.23 |
ENSMUST00000054920.5
|
Myorg
|
myogenesis regulating glycosidase (putative) |
| chr10_+_116137277 | 1.22 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr10_-_128755127 | 1.22 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr2_+_130748380 | 1.22 |
ENSMUST00000028781.9
|
Atrn
|
attractin |
| chr7_+_140500808 | 1.21 |
ENSMUST00000106045.8
ENSMUST00000183845.8 |
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr1_-_189902868 | 1.21 |
ENSMUST00000177288.4
ENSMUST00000175916.8 |
Prox1
|
prospero homeobox 1 |
| chr4_+_137640782 | 1.19 |
ENSMUST00000151110.4
|
Ece1
|
endothelin converting enzyme 1 |
| chr9_-_107556823 | 1.18 |
ENSMUST00000010205.9
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr9_-_56325344 | 1.18 |
ENSMUST00000061552.15
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr3_-_116762476 | 1.18 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr19_+_23118545 | 1.18 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9 |
| chr18_-_46861414 | 1.17 |
ENSMUST00000234819.2
ENSMUST00000035804.9 |
Cdo1
|
cysteine dioxygenase 1, cytosolic |
| chr12_-_103871146 | 1.16 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr14_-_21102188 | 1.15 |
ENSMUST00000130370.3
ENSMUST00000224016.2 ENSMUST00000022371.10 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr14_+_21102642 | 1.14 |
ENSMUST00000045376.11
|
Adk
|
adenosine kinase |
| chr16_-_46317135 | 1.13 |
ENSMUST00000149901.2
ENSMUST00000096052.9 |
Nectin3
|
nectin cell adhesion molecule 3 |
| chr3_-_75864195 | 1.13 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr13_-_55574582 | 1.13 |
ENSMUST00000170921.2
|
F12
|
coagulation factor XII (Hageman factor) |
| chr19_-_7779943 | 1.11 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr11_-_70590923 | 1.10 |
ENSMUST00000108543.4
ENSMUST00000108542.8 ENSMUST00000108541.9 ENSMUST00000126114.9 ENSMUST00000073625.8 |
Inca1
|
inhibitor of CDK, cyclin A1 interacting protein 1 |
| chr9_-_103105638 | 1.10 |
ENSMUST00000126359.2
|
Trf
|
transferrin |
| chr11_+_84070593 | 1.08 |
ENSMUST00000137500.9
ENSMUST00000130012.9 |
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr17_+_64170967 | 1.07 |
ENSMUST00000232945.2
|
Fer
|
fer (fms/fps related) protein kinase |
| chr4_+_140970161 | 1.05 |
ENSMUST00000138096.8
ENSMUST00000006618.9 ENSMUST00000125392.8 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr14_+_20732804 | 1.05 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr3_+_94284812 | 1.04 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr1_+_133173826 | 1.01 |
ENSMUST00000105082.9
ENSMUST00000038295.15 |
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr17_-_56428968 | 1.01 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr9_-_44632680 | 1.01 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr1_-_134163102 | 0.99 |
ENSMUST00000187631.2
ENSMUST00000038191.8 ENSMUST00000086465.6 |
Adora1
|
adenosine A1 receptor |
| chr6_+_34722926 | 0.99 |
ENSMUST00000126181.8
|
Cald1
|
caldesmon 1 |
| chrX_-_51254129 | 0.98 |
ENSMUST00000033450.3
|
Gpc4
|
glypican 4 |
| chr11_+_114566257 | 0.97 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr6_+_134807170 | 0.97 |
ENSMUST00000111937.2
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chrX_-_94209913 | 0.96 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr11_+_75084609 | 0.96 |
ENSMUST00000102514.4
|
Rtn4rl1
|
reticulon 4 receptor-like 1 |
| chr1_-_180823709 | 0.95 |
ENSMUST00000154133.8
|
Ephx1
|
epoxide hydrolase 1, microsomal |
| chr11_+_98932062 | 0.95 |
ENSMUST00000017637.13
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr12_-_103829810 | 0.95 |
ENSMUST00000085056.8
ENSMUST00000072876.12 ENSMUST00000124717.2 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr18_-_34639848 | 0.94 |
ENSMUST00000040506.8
|
Fam13b
|
family with sequence similarity 13, member B |
| chr18_+_84106796 | 0.94 |
ENSMUST00000235383.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr2_+_33106062 | 0.93 |
ENSMUST00000004208.7
|
Angptl2
|
angiopoietin-like 2 |
| chr11_-_4068779 | 0.93 |
ENSMUST00000003681.8
|
Sec14l2
|
SEC14-like lipid binding 2 |
| chr12_-_81014755 | 0.93 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr10_+_98943999 | 0.92 |
ENSMUST00000161240.4
|
Galnt4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr11_+_80044931 | 0.92 |
ENSMUST00000021050.14
|
Adap2
|
ArfGAP with dual PH domains 2 |
| chr2_+_24944367 | 0.92 |
ENSMUST00000100334.11
ENSMUST00000152122.8 ENSMUST00000116574.10 ENSMUST00000006646.15 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr7_+_127070180 | 0.90 |
ENSMUST00000133817.8
ENSMUST00000133938.8 |
Prr14
|
proline rich 14 |
| chr5_+_89175894 | 0.90 |
ENSMUST00000113216.9
ENSMUST00000134303.2 |
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr9_-_71678814 | 0.90 |
ENSMUST00000122065.2
ENSMUST00000121322.8 ENSMUST00000072899.9 |
Cgnl1
|
cingulin-like 1 |
| chr10_+_87697155 | 0.90 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr7_+_140415170 | 0.90 |
ENSMUST00000211372.2
ENSMUST00000026554.11 ENSMUST00000185612.3 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr4_-_53159885 | 0.90 |
ENSMUST00000030010.4
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr6_+_17463925 | 0.89 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr7_-_126483851 | 0.88 |
ENSMUST00000071268.11
ENSMUST00000117394.2 |
Taok2
|
TAO kinase 2 |
| chr3_+_89136353 | 0.88 |
ENSMUST00000041142.4
|
Muc1
|
mucin 1, transmembrane |
| chr14_-_26256025 | 0.87 |
ENSMUST00000139075.8
ENSMUST00000102956.8 |
Slmap
|
sarcolemma associated protein |
| chr16_-_45830575 | 0.87 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr1_-_72914036 | 0.87 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr1_-_172125555 | 0.86 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr5_-_116560916 | 0.86 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr6_+_121320339 | 0.86 |
ENSMUST00000168295.2
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr1_+_62742444 | 0.86 |
ENSMUST00000102822.9
ENSMUST00000075144.12 |
Nrp2
|
neuropilin 2 |
| chr2_+_160573178 | 0.85 |
ENSMUST00000103115.8
|
Plcg1
|
phospholipase C, gamma 1 |
| chr14_+_34097422 | 0.85 |
ENSMUST00000111908.3
|
Mmrn2
|
multimerin 2 |
| chr7_+_26508305 | 0.84 |
ENSMUST00000040944.9
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr11_-_53782462 | 0.84 |
ENSMUST00000019044.8
|
Slc22a5
|
solute carrier family 22 (organic cation transporter), member 5 |
| chr4_-_41569500 | 0.84 |
ENSMUST00000108049.9
ENSMUST00000108052.10 ENSMUST00000108050.2 |
Fam219a
|
family with sequence similarity 219, member A |
| chr15_+_54975713 | 0.83 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr10_-_31321793 | 0.83 |
ENSMUST00000213639.2
ENSMUST00000215515.2 ENSMUST00000214644.2 ENSMUST00000213528.2 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr1_-_121255400 | 0.82 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr12_-_75642969 | 0.82 |
ENSMUST00000021447.9
ENSMUST00000220035.2 |
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr1_-_65225572 | 0.82 |
ENSMUST00000188109.7
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr11_-_77784922 | 0.82 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr16_+_35803794 | 0.81 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr12_-_81014849 | 0.80 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr10_+_75729237 | 0.80 |
ENSMUST00000009236.6
ENSMUST00000217811.2 |
Derl3
|
Der1-like domain family, member 3 |
| chr7_-_116042674 | 0.80 |
ENSMUST00000170430.3
ENSMUST00000206219.2 |
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha |
| chr17_+_34482183 | 0.79 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr1_-_98023321 | 0.79 |
ENSMUST00000058762.15
ENSMUST00000097625.10 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_33106121 | 0.79 |
ENSMUST00000193373.3
|
Angptl2
|
angiopoietin-like 2 |
| chr16_-_23339329 | 0.78 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr12_+_78273356 | 0.78 |
ENSMUST00000110388.10
|
Gphn
|
gephyrin |
| chr11_-_107607343 | 0.77 |
ENSMUST00000021065.6
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr10_+_4660119 | 0.77 |
ENSMUST00000105588.8
ENSMUST00000105589.2 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr15_+_54975814 | 0.77 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr13_-_104246084 | 0.75 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr2_-_118377500 | 0.75 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor |
| chr9_-_103099262 | 0.75 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr12_+_73954678 | 0.75 |
ENSMUST00000110464.8
ENSMUST00000021530.8 |
Hif1a
|
hypoxia inducible factor 1, alpha subunit |
| chr8_-_49008305 | 0.75 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_157240138 | 0.74 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr11_-_88742285 | 0.74 |
ENSMUST00000107903.8
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr14_-_65187287 | 0.74 |
ENSMUST00000067843.10
ENSMUST00000176489.8 ENSMUST00000175905.8 ENSMUST00000022544.14 ENSMUST00000175744.8 ENSMUST00000176128.8 |
Hmbox1
|
homeobox containing 1 |
| chr7_+_89053562 | 0.74 |
ENSMUST00000058755.5
|
Fzd4
|
frizzled class receptor 4 |
| chr16_-_23339548 | 0.73 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr14_+_34097474 | 0.73 |
ENSMUST00000227130.2
|
Mmrn2
|
multimerin 2 |
| chr5_+_52521133 | 0.73 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr14_-_70445086 | 0.73 |
ENSMUST00000022682.6
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr11_+_48728291 | 0.73 |
ENSMUST00000046903.6
|
Trim7
|
tripartite motif-containing 7 |
| chr2_-_18397547 | 0.73 |
ENSMUST00000091418.12
ENSMUST00000166495.8 |
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr7_+_100355798 | 0.73 |
ENSMUST00000107042.9
ENSMUST00000207564.2 ENSMUST00000049053.9 |
Fam168a
|
family with sequence similarity 168, member A |
| chr1_+_194302123 | 0.73 |
ENSMUST00000027952.12
|
Plxna2
|
plexin A2 |
| chr17_-_26417982 | 0.72 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr11_-_59927688 | 0.72 |
ENSMUST00000102692.10
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr7_+_51528715 | 0.72 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr11_+_35660288 | 0.72 |
ENSMUST00000018990.8
|
Pank3
|
pantothenate kinase 3 |
| chr18_+_84106188 | 0.72 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr14_-_70588803 | 0.72 |
ENSMUST00000143153.2
ENSMUST00000127000.2 ENSMUST00000068044.14 ENSMUST00000022688.10 |
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr7_-_126873219 | 0.71 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr10_+_127595590 | 0.69 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr2_-_179915276 | 0.69 |
ENSMUST00000108891.2
|
Cables2
|
CDK5 and Abl enzyme substrate 2 |
| chr15_-_78352801 | 0.69 |
ENSMUST00000229124.2
ENSMUST00000230226.2 ENSMUST00000017086.5 |
Tmprss6
|
transmembrane serine protease 6 |
| chr11_+_53241561 | 0.69 |
ENSMUST00000060945.12
|
Aff4
|
AF4/FMR2 family, member 4 |
| chr5_+_119808722 | 0.69 |
ENSMUST00000079719.11
|
Tbx3
|
T-box 3 |
| chr14_-_72946972 | 0.67 |
ENSMUST00000162478.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr2_+_20742115 | 0.67 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr9_+_51959534 | 0.66 |
ENSMUST00000061352.11
|
Rdx
|
radixin |
| chr11_-_4110286 | 0.66 |
ENSMUST00000093381.11
ENSMUST00000101626.9 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr10_-_128425519 | 0.66 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr14_+_54669054 | 0.66 |
ENSMUST00000089688.6
ENSMUST00000225641.2 |
Mmp14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr3_+_95226093 | 0.66 |
ENSMUST00000139866.2
|
Cers2
|
ceramide synthase 2 |
| chr11_+_69983531 | 0.66 |
ENSMUST00000124721.2
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr7_+_140415431 | 0.66 |
ENSMUST00000209978.2
ENSMUST00000210916.2 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr6_+_90442269 | 0.66 |
ENSMUST00000113530.4
|
Klf15
|
Kruppel-like factor 15 |
| chr11_+_119493806 | 0.66 |
ENSMUST00000026671.13
|
Rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr11_+_75358866 | 0.65 |
ENSMUST00000043598.14
ENSMUST00000108435.2 |
Tlcd2
|
TLC domain containing 2 |
| chr9_-_110576124 | 0.65 |
ENSMUST00000199862.5
ENSMUST00000198865.5 |
Pth1r
|
parathyroid hormone 1 receptor |
| chr8_+_119010458 | 0.65 |
ENSMUST00000117160.2
|
Cdh13
|
cadherin 13 |
| chr8_+_94763826 | 0.65 |
ENSMUST00000109556.9
ENSMUST00000093301.9 ENSMUST00000060632.8 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr15_+_6599001 | 0.65 |
ENSMUST00000227175.2
|
Fyb
|
FYN binding protein |
| chr8_-_106670014 | 0.65 |
ENSMUST00000038896.8
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr3_+_135191366 | 0.65 |
ENSMUST00000029814.10
|
Manba
|
mannosidase, beta A, lysosomal |
| chr17_+_24022153 | 0.65 |
ENSMUST00000190686.7
ENSMUST00000088621.11 ENSMUST00000233636.2 |
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr13_+_38335232 | 0.64 |
ENSMUST00000124830.3
|
Dsp
|
desmoplakin |
| chr7_+_125871761 | 0.64 |
ENSMUST00000056028.11
|
Sbk1
|
SH3-binding kinase 1 |
| chr1_-_55127312 | 0.64 |
ENSMUST00000127861.8
ENSMUST00000144077.3 |
Hspd1
|
heat shock protein 1 (chaperonin) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 1.4 | 5.5 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.9 | 2.6 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.8 | 2.5 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.7 | 2.9 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.7 | 6.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.6 | 2.5 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.6 | 3.7 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.6 | 1.8 | GO:0003167 | atrioventricular bundle cell differentiation(GO:0003167) |
| 0.5 | 1.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.5 | 1.8 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.4 | 2.6 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.4 | 1.2 | GO:0006867 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 0.4 | 1.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.4 | 1.2 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 1.0 | GO:0032242 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.3 | 1.5 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.3 | 0.6 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.3 | 0.8 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.3 | 0.8 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.3 | 1.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 0.8 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.3 | 0.8 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.3 | 1.3 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.3 | 3.3 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.3 | 0.8 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.2 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 0.7 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 8.3 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 | 0.9 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.2 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 0.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 1.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.2 | 1.1 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.2 | 0.7 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 1.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 0.9 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.2 | 0.9 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.2 | 3.0 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 1.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.6 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 0.6 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.2 | 1.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 4.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 1.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 1.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 0.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 1.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.2 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 1.5 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 2.9 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 0.5 | GO:1904761 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) negative regulation of myofibroblast differentiation(GO:1904761) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.2 | 0.7 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 0.9 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.2 | 1.1 | GO:0019452 | L-cysteine catabolic process to taurine(GO:0019452) |
| 0.2 | 0.7 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.2 | 0.9 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.2 | 1.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.7 | GO:0046226 | coumarin metabolic process(GO:0009804) coumarin catabolic process(GO:0046226) |
| 0.2 | 0.6 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 1.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.5 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.2 | 2.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.2 | 1.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.2 | 2.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.6 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.4 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 2.5 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.1 | 0.3 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.1 | 0.4 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.1 | 1.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 2.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 1.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.5 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.6 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.6 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.9 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.9 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.4 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.4 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.5 | GO:0050822 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 0.4 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 2.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.5 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.8 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.6 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.7 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 | 0.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.7 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.8 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.6 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 0.3 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.7 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.6 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.4 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.8 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 2.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.2 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.7 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.1 | 2.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 1.6 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.4 | GO:1902045 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 1.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.5 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 1.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.3 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 2.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 1.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.7 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.6 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 0.4 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 1.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.5 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 1.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.8 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 4.4 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.1 | 0.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.7 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.6 | GO:0048597 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 1.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.6 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.1 | 1.0 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 1.6 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.6 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.9 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.1 | 0.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 1.3 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.7 | GO:0048087 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of developmental pigmentation(GO:0048087) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 0.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.6 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.8 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.6 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 1.5 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.9 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 1.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.5 | GO:0043482 | cellular pigment accumulation(GO:0043482) |
| 0.1 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 4.4 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 1.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 1.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 3.6 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.1 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 2.9 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.8 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 1.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.3 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in kidney development(GO:0035787) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 3.9 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 1.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 1.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 3.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0003256 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.0 | 0.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.6 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:0071372 | response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 1.0 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0002362 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 1.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.0 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.0 | 0.1 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 1.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.2 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.7 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0016045 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 0.5 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.3 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.0 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.6 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.0 | 0.1 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.7 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.3 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) paranodal junction assembly(GO:0030913) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.7 | GO:0090314 | positive regulation of protein targeting to membrane(GO:0090314) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0002001 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) regulation of renin secretion into blood stream(GO:1900133) positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.6 | 2.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.5 | 1.6 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.4 | 3.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 1.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 5.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 5.9 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 2.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 1.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 4.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 2.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.4 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.4 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.1 | 2.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 3.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 0.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 1.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 4.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 4.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 7.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 7.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 3.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 4.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 19.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.5 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 1.0 | 4.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.0 | 3.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.9 | 5.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.9 | 2.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.6 | 3.7 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.6 | 2.9 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.5 | 1.6 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.5 | 1.8 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.4 | 10.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 1.7 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.4 | 2.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 2.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.4 | 1.1 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 3.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.0 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 1.2 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.3 | 1.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 1.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 1.5 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.3 | 2.0 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.3 | 0.9 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.3 | 2.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.3 | 1.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 1.8 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.7 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.2 | 1.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 1.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 1.5 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.2 | 0.7 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.2 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.6 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.2 | 0.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 0.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 0.6 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 2.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.4 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.2 | 0.7 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 8.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 0.7 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 1.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 4.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.7 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.2 | 1.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 0.8 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 0.9 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 5.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.8 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 1.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.4 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 1.3 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 3.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 2.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 4.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.6 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.3 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.8 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 4.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.9 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.1 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.5 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.9 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 1.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 1.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.4 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 2.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 1.4 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 1.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.6 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 1.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 3.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 4.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 1.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.9 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 9.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 3.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 9.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 8.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 2.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 4.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 5.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 1.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 4.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.9 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |