Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
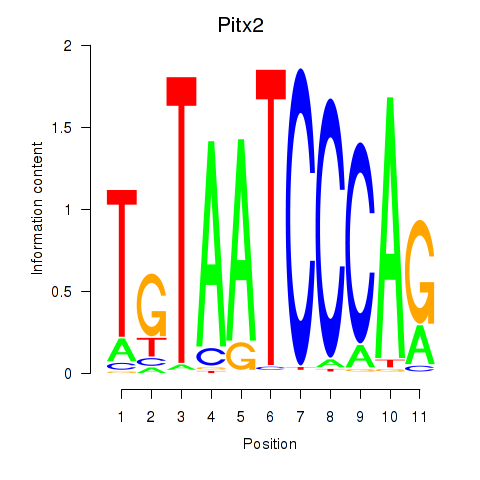
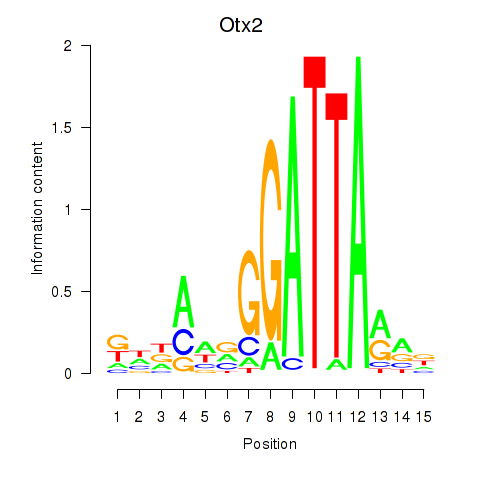
Results for Pitx2_Otx2
Z-value: 3.46
Transcription factors associated with Pitx2_Otx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx2
|
ENSMUSG00000028023.17 | Pitx2 |
|
Otx2
|
ENSMUSG00000021848.17 | Otx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx2 | mm39_v1_chr3_+_129007599_129007621 | 0.66 | 1.2e-05 | Click! |
| Otx2 | mm39_v1_chr14_-_48902555_48902572 | 0.33 | 4.8e-02 | Click! |
Activity profile of Pitx2_Otx2 motif
Sorted Z-values of Pitx2_Otx2 motif
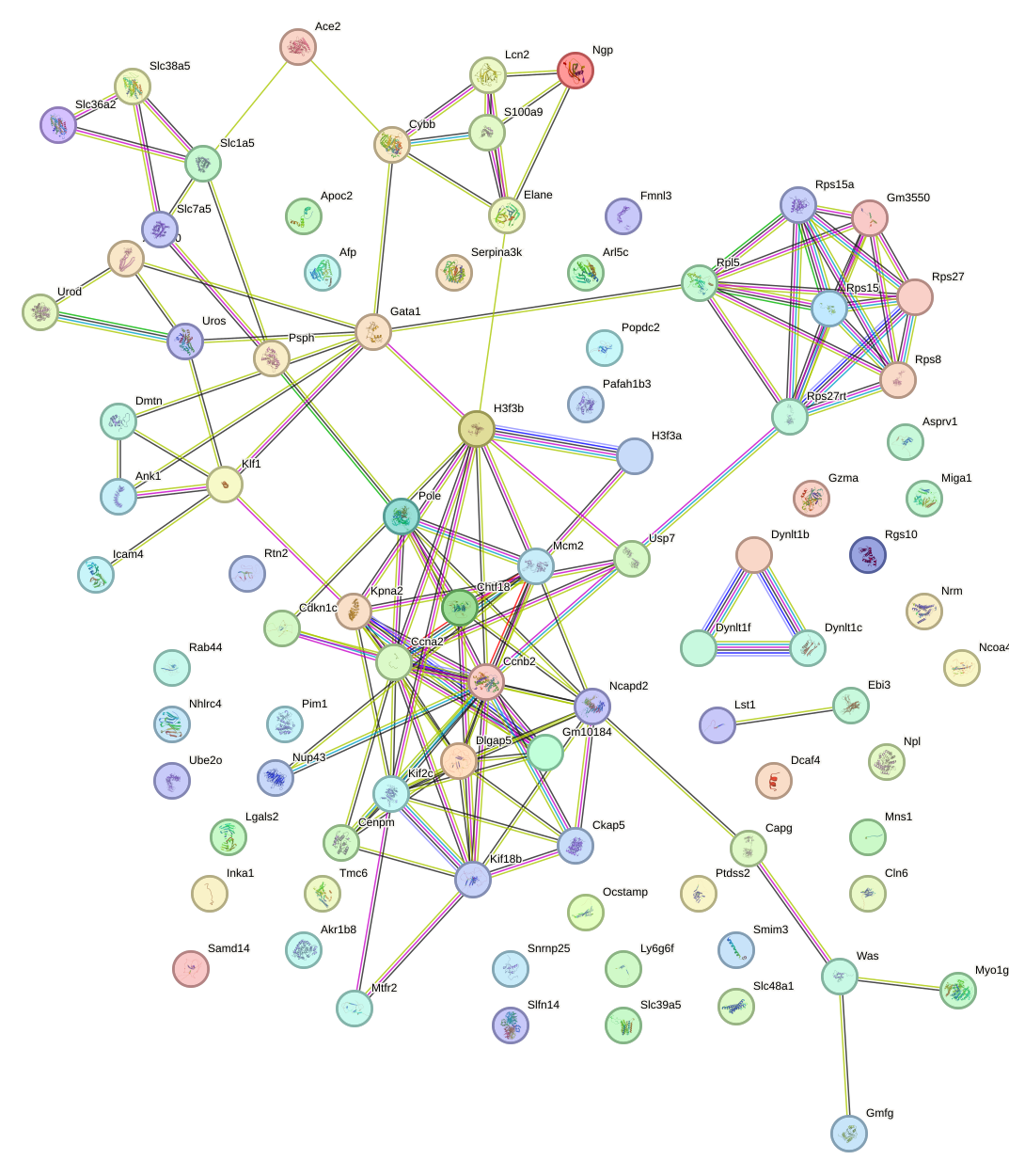
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx2_Otx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110248815 | 36.30 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr16_+_36004452 | 35.63 |
ENSMUST00000114858.2
|
Cstdc4
|
cystatin domain containing 4 |
| chr5_+_90638580 | 27.28 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr16_-_36188086 | 26.46 |
ENSMUST00000096089.3
|
Cstdc5
|
cystatin domain containing 5 |
| chr8_+_85628557 | 22.04 |
ENSMUST00000067060.10
ENSMUST00000239392.2 |
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chr10_+_79722081 | 20.70 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr10_+_128736827 | 19.50 |
ENSMUST00000219317.2
|
Cd63
|
CD63 antigen |
| chr6_-_88875646 | 18.08 |
ENSMUST00000058011.8
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr3_-_90603013 | 17.38 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr16_+_36097313 | 17.33 |
ENSMUST00000232150.2
|
Stfa1
|
stefin A1 |
| chr17_-_35407403 | 17.33 |
ENSMUST00000097336.5
|
Lst1
|
leukocyte specific transcript 1 |
| chr17_+_36172210 | 17.20 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr11_-_6470918 | 15.89 |
ENSMUST00000003459.4
|
Myo1g
|
myosin IG |
| chr14_-_70867588 | 15.58 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr7_+_16515265 | 15.57 |
ENSMUST00000108496.9
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr10_-_62178453 | 14.61 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr15_-_78739717 | 14.36 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr7_-_45173193 | 13.26 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr2_+_91376650 | 12.94 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr3_-_36626101 | 12.52 |
ENSMUST00000029270.10
|
Ccna2
|
cyclin A2 |
| chr2_-_32278245 | 12.05 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr11_-_97886997 | 11.84 |
ENSMUST00000042971.16
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr7_-_45175570 | 11.76 |
ENSMUST00000167273.2
ENSMUST00000042105.11 |
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr2_-_165242307 | 11.58 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr7_+_140711181 | 11.58 |
ENSMUST00000026568.10
|
Ptdss2
|
phosphatidylserine synthase 2 |
| chr7_-_143014726 | 11.53 |
ENSMUST00000167912.9
ENSMUST00000037287.8 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr14_+_31888061 | 11.47 |
ENSMUST00000164341.2
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chr7_-_143013899 | 11.45 |
ENSMUST00000208137.2
ENSMUST00000207910.2 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr16_+_36097505 | 11.39 |
ENSMUST00000042097.11
|
Stfa1
|
stefin A1 |
| chr11_+_61847589 | 11.36 |
ENSMUST00000201671.4
ENSMUST00000202178.4 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr9_-_70328816 | 11.09 |
ENSMUST00000034742.8
|
Ccnb2
|
cyclin B2 |
| chr6_-_115739284 | 11.04 |
ENSMUST00000166254.7
ENSMUST00000170625.8 |
Tmem40
|
transmembrane protein 40 |
| chr12_+_83572774 | 11.03 |
ENSMUST00000223291.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr11_-_116472272 | 10.82 |
ENSMUST00000082152.5
|
Ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr10_-_128237087 | 10.78 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr11_-_83177548 | 10.73 |
ENSMUST00000163961.3
|
Slfn14
|
schlafen 14 |
| chr17_-_26163906 | 10.68 |
ENSMUST00000085027.4
ENSMUST00000237570.2 |
Nhlrc4
Pigq
|
NHL repeat containing 4 phosphatidylinositol glycan anchor biosynthesis, class Q |
| chrX_+_8137881 | 10.36 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_-_7844265 | 10.22 |
ENSMUST00000125418.2
|
Gata1
|
GATA binding protein 1 |
| chr4_-_117039809 | 10.17 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr11_+_117673107 | 10.07 |
ENSMUST00000050874.14
ENSMUST00000106334.9 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr17_-_35304582 | 9.92 |
ENSMUST00000038507.7
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr17_+_36172235 | 9.63 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr10_+_80128248 | 9.62 |
ENSMUST00000068408.14
ENSMUST00000062674.7 |
Rps15
|
ribosomal protein S15 |
| chr9_+_20940669 | 9.61 |
ENSMUST00000001040.7
ENSMUST00000215077.2 |
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr17_+_29712008 | 9.56 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chr18_-_60635059 | 9.40 |
ENSMUST00000042710.8
|
Smim3
|
small integral membrane protein 3 |
| chr17_-_6923299 | 9.32 |
ENSMUST00000179554.3
|
Dynlt1f
|
dynein light chain Tctex-type 1F |
| chr9_+_72345267 | 9.22 |
ENSMUST00000183809.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr11_+_94901104 | 9.10 |
ENSMUST00000124735.2
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr11_-_106889291 | 9.09 |
ENSMUST00000124541.8
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr15_+_97682210 | 9.09 |
ENSMUST00000117892.2
ENSMUST00000229084.2 |
Slc48a1
|
solute carrier family 48 (heme transporter), member 1 |
| chr9_-_107863062 | 8.81 |
ENSMUST00000048568.6
|
Inka1
|
inka box actin regulator 1 |
| chr7_+_28140450 | 8.75 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr17_-_33932346 | 8.63 |
ENSMUST00000173392.2
|
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr11_+_61847622 | 8.61 |
ENSMUST00000202389.4
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr10_+_128744689 | 8.50 |
ENSMUST00000105229.9
|
Cd63
|
CD63 antigen |
| chr6_-_125168637 | 8.49 |
ENSMUST00000043848.11
|
Ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr1_-_153425791 | 8.19 |
ENSMUST00000041874.9
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr11_-_55075855 | 8.13 |
ENSMUST00000039305.6
|
Slc36a2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr17_+_6697511 | 8.08 |
ENSMUST00000179569.3
|
Dynlt1b
|
dynein light chain Tctex-type 1B |
| chr16_+_38182569 | 7.96 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr7_-_19410749 | 7.91 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr4_-_117013396 | 7.82 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
| chr18_+_34869412 | 7.73 |
ENSMUST00000105038.3
|
Gm3550
|
predicted gene 3550 |
| chr16_-_36154692 | 7.67 |
ENSMUST00000114850.3
|
Cstdc6
|
cystatin domain containing 6 |
| chrX_+_162923474 | 7.65 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr13_-_113237505 | 7.64 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr7_+_28140352 | 7.47 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chrX_-_9335525 | 7.45 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr15_-_82128888 | 7.43 |
ENSMUST00000089155.6
ENSMUST00000089157.11 |
Cenpm
|
centromere protein M |
| chr14_-_47655621 | 7.41 |
ENSMUST00000180299.8
|
Dlgap5
|
DLG associated protein 5 |
| chr5_-_100720063 | 7.32 |
ENSMUST00000031264.12
|
Plac8
|
placenta-specific 8 |
| chr8_-_124709859 | 7.21 |
ENSMUST00000075578.7
|
Abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr6_+_72521374 | 7.19 |
ENSMUST00000071044.13
ENSMUST00000114072.8 |
Capg
|
capping protein (actin filament), gelsolin-like |
| chr17_-_25946370 | 7.11 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr8_-_122634418 | 7.07 |
ENSMUST00000045557.10
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr16_+_36030773 | 7.04 |
ENSMUST00000089628.4
|
Csta3
|
cystatin A family member 3 |
| chr7_-_24997291 | 7.00 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr11_+_32155415 | 6.97 |
ENSMUST00000039601.10
ENSMUST00000149043.3 |
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr10_+_20223516 | 6.94 |
ENSMUST00000169712.3
ENSMUST00000217608.2 |
Mtfr2
|
mitochondrial fission regulator 2 |
| chr3_-_152045986 | 6.92 |
ENSMUST00000199397.2
ENSMUST00000199334.5 ENSMUST00000068243.11 ENSMUST00000073089.13 |
Miga1
|
mitoguardin 1 |
| chr7_-_128019874 | 6.84 |
ENSMUST00000145739.3
ENSMUST00000033133.12 |
Rgs10
|
regulator of G-protein signalling 10 |
| chr8_+_23629080 | 6.82 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr6_+_86605146 | 6.74 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr7_-_117715394 | 6.59 |
ENSMUST00000131374.8
|
Rps15a
|
ribosomal protein S15A |
| chr16_-_8610165 | 6.57 |
ENSMUST00000160405.8
|
Usp7
|
ubiquitin specific peptidase 7 |
| chr11_+_114559350 | 6.55 |
ENSMUST00000106602.10
ENSMUST00000077915.10 ENSMUST00000106599.8 ENSMUST00000082092.5 |
Rpl38
|
ribosomal protein L38 |
| chr6_+_34331054 | 6.52 |
ENSMUST00000038406.7
|
Akr1b8
|
aldo-keto reductase family 1, member B8 |
| chr5_+_110478558 | 6.48 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr17_+_56259617 | 6.47 |
ENSMUST00000003274.8
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr9_+_72345801 | 6.40 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr4_-_116851550 | 6.37 |
ENSMUST00000130273.8
|
Urod
|
uroporphyrinogen decarboxylase |
| chr7_-_133304244 | 6.36 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chrX_-_7956682 | 6.30 |
ENSMUST00000033505.7
|
Was
|
Wiskott-Aldrich syndrome |
| chr17_+_29333116 | 6.28 |
ENSMUST00000233717.2
ENSMUST00000141239.2 |
Rab44
|
RAB44, member RAS oncogene family |
| chr5_-_129856237 | 6.28 |
ENSMUST00000118268.9
|
Psph
|
phosphoserine phosphatase |
| chr11_-_102815910 | 6.14 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr11_-_115918784 | 6.10 |
ENSMUST00000106454.8
|
H3f3b
|
H3.3 histone B |
| chr10_+_7543260 | 6.09 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr5_-_129856267 | 6.03 |
ENSMUST00000201394.4
|
Psph
|
phosphoserine phosphatase |
| chr4_-_116851571 | 5.99 |
ENSMUST00000030446.15
|
Urod
|
uroporphyrinogen decarboxylase |
| chr9_-_114811807 | 5.81 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr15_-_99268311 | 5.79 |
ENSMUST00000081224.14
ENSMUST00000120633.2 ENSMUST00000088233.13 |
Fmnl3
|
formin-like 3 |
| chr9_+_62754252 | 5.73 |
ENSMUST00000124984.2
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr7_-_100164007 | 5.70 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr5_+_108048367 | 5.69 |
ENSMUST00000082223.13
|
Rpl5
|
ribosomal protein L5 |
| chr11_-_117673008 | 5.64 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr7_+_19024994 | 5.60 |
ENSMUST00000108468.5
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr4_-_149221998 | 5.59 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr19_-_6996791 | 5.54 |
ENSMUST00000040772.9
|
Fermt3
|
fermitin family member 3 |
| chr15_-_76554065 | 5.51 |
ENSMUST00000037824.6
|
Foxh1
|
forkhead box H1 |
| chr17_+_48539782 | 5.50 |
ENSMUST00000113251.10
ENSMUST00000048782.7 |
Trem1
|
triggering receptor expressed on myeloid cells 1 |
| chr19_+_4204605 | 5.49 |
ENSMUST00000061086.9
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr7_-_24997393 | 5.41 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr9_-_58648826 | 5.38 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr19_-_17316906 | 5.38 |
ENSMUST00000169897.2
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr9_+_103182352 | 5.34 |
ENSMUST00000035164.10
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr7_-_110581652 | 5.33 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr4_-_149222057 | 5.30 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr3_-_105839980 | 5.29 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr19_+_8861096 | 5.28 |
ENSMUST00000187504.7
|
Lbhd1
|
LBH domain containing 1 |
| chr6_+_35154319 | 5.27 |
ENSMUST00000201374.4
ENSMUST00000043815.16 |
Nup205
|
nucleoporin 205 |
| chr3_+_129672205 | 5.19 |
ENSMUST00000029629.15
|
Pla2g12a
|
phospholipase A2, group XIIA |
| chr8_+_23629046 | 5.17 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr11_+_68936457 | 5.11 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chrX_-_161747552 | 5.08 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr14_+_71011744 | 5.06 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr11_+_101333115 | 5.03 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr19_+_8870362 | 5.02 |
ENSMUST00000096249.7
|
Ints5
|
integrator complex subunit 5 |
| chr4_+_154096188 | 5.00 |
ENSMUST00000169622.8
ENSMUST00000030894.15 |
Lrrc47
|
leucine rich repeat containing 47 |
| chr17_+_6869070 | 4.97 |
ENSMUST00000092966.5
|
Dynlt1c
|
dynein light chain Tctex-type 1C |
| chr11_-_101979297 | 4.93 |
ENSMUST00000017458.11
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr19_+_37364791 | 4.90 |
ENSMUST00000012587.4
|
Kif11
|
kinesin family member 11 |
| chr15_-_89310060 | 4.87 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr14_-_54949596 | 4.86 |
ENSMUST00000064290.8
|
Cebpe
|
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr7_+_141027763 | 4.83 |
ENSMUST00000106003.2
|
Rplp2
|
ribosomal protein, large P2 |
| chr8_+_118225008 | 4.81 |
ENSMUST00000081232.9
|
Plcg2
|
phospholipase C, gamma 2 |
| chr14_+_66043515 | 4.81 |
ENSMUST00000139644.2
|
Pbk
|
PDZ binding kinase |
| chr6_-_131270136 | 4.79 |
ENSMUST00000032307.12
|
Magohb
|
mago homolog B, exon junction complex core component |
| chr7_-_30144933 | 4.74 |
ENSMUST00000006828.9
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr15_+_59186876 | 4.65 |
ENSMUST00000022977.14
ENSMUST00000100640.5 |
Sqle
|
squalene epoxidase |
| chr15_-_83479312 | 4.62 |
ENSMUST00000016901.5
|
Ttll12
|
tubulin tyrosine ligase-like family, member 12 |
| chr5_-_139722046 | 4.61 |
ENSMUST00000044642.14
|
Micall2
|
MICAL-like 2 |
| chr10_+_62782786 | 4.57 |
ENSMUST00000131422.8
|
Dna2
|
DNA replication helicase/nuclease 2 |
| chr2_-_126342551 | 4.56 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr4_-_156340276 | 4.54 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr1_-_128520002 | 4.51 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr13_-_97235745 | 4.48 |
ENSMUST00000071118.7
|
Gm6169
|
predicted gene 6169 |
| chr7_-_30119227 | 4.48 |
ENSMUST00000208740.2
ENSMUST00000075062.5 |
Hcst
|
hematopoietic cell signal transducer |
| chr15_+_80832685 | 4.45 |
ENSMUST00000023043.10
ENSMUST00000164806.6 ENSMUST00000207170.2 ENSMUST00000168756.8 |
Adsl
|
adenylosuccinate lyase |
| chr2_+_32477069 | 4.43 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr8_-_11685726 | 4.40 |
ENSMUST00000033905.13
ENSMUST00000169782.3 |
Ankrd10
|
ankyrin repeat domain 10 |
| chr2_-_5900130 | 4.40 |
ENSMUST00000026926.5
ENSMUST00000193792.6 ENSMUST00000102981.10 |
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae) |
| chr10_+_128073900 | 4.38 |
ENSMUST00000105245.3
|
Timeless
|
timeless circadian clock 1 |
| chr5_+_115697526 | 4.32 |
ENSMUST00000086519.12
ENSMUST00000156359.2 ENSMUST00000152976.2 |
Rplp0
|
ribosomal protein, large, P0 |
| chr11_-_5102218 | 4.29 |
ENSMUST00000163299.8
ENSMUST00000062821.13 |
Emid1
|
EMI domain containing 1 |
| chr7_+_127503251 | 4.28 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr14_+_43951187 | 4.28 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr1_-_172722589 | 4.26 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr12_-_69205882 | 4.22 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr8_+_61343444 | 4.21 |
ENSMUST00000146863.2
ENSMUST00000037190.15 |
Hpf1
|
histone PARylation factor 1 |
| chr10_-_79875479 | 4.17 |
ENSMUST00000004786.10
|
Polr2e
|
polymerase (RNA) II (DNA directed) polypeptide E |
| chr7_-_47178610 | 4.14 |
ENSMUST00000172559.2
|
Mrgpra2b
|
MAS-related GPR, member A2B |
| chr11_-_84719779 | 4.12 |
ENSMUST00000047560.8
|
Dhrs11
|
dehydrogenase/reductase (SDR family) member 11 |
| chr12_-_75678092 | 4.12 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr2_+_174257597 | 4.11 |
ENSMUST00000109075.8
ENSMUST00000016397.7 |
Nelfcd
|
negative elongation factor complex member C/D, Th1l |
| chr2_-_34645241 | 4.11 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr4_+_156194427 | 4.09 |
ENSMUST00000072554.13
ENSMUST00000169550.8 ENSMUST00000105576.2 |
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene |
| chr14_-_54655079 | 4.07 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr5_+_139239247 | 4.07 |
ENSMUST00000138508.8
ENSMUST00000110878.2 |
Get4
|
golgi to ER traffic protein 4 |
| chr10_+_79650496 | 4.01 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr9_+_20799471 | 3.99 |
ENSMUST00000004203.6
|
Ppan
|
peter pan homolog |
| chr7_+_101530524 | 3.98 |
ENSMUST00000106978.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr14_-_99231754 | 3.97 |
ENSMUST00000081987.5
|
Rpl36a-ps1
|
ribosomal protein L36A, pseudogene 1 |
| chr15_-_36598263 | 3.97 |
ENSMUST00000155116.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr18_+_35932894 | 3.95 |
ENSMUST00000236484.2
ENSMUST00000236550.2 ENSMUST00000235919.2 |
Ube2d2a
Gm50371
|
ubiquitin-conjugating enzyme E2D 2A predicted gene, 50371 |
| chr11_-_120238917 | 3.95 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr15_-_82128538 | 3.93 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr14_-_70414236 | 3.92 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr9_+_119939414 | 3.92 |
ENSMUST00000035106.12
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr1_-_23948764 | 3.91 |
ENSMUST00000129254.8
|
Smap1
|
small ArfGAP 1 |
| chr4_+_130640436 | 3.89 |
ENSMUST00000151698.8
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr12_-_79027531 | 3.88 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr11_-_78074377 | 3.86 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr10_+_79832313 | 3.83 |
ENSMUST00000132517.8
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr16_-_4536943 | 3.82 |
ENSMUST00000120056.8
ENSMUST00000074970.8 |
Nmral1
|
NmrA-like family domain containing 1 |
| chr7_+_16726633 | 3.79 |
ENSMUST00000094805.5
|
Ccdc8
|
coiled-coil domain containing 8 |
| chr1_-_167112784 | 3.77 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr16_+_55786638 | 3.77 |
ENSMUST00000023269.5
|
Rpl24
|
ribosomal protein L24 |
| chr18_-_74340842 | 3.74 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr16_+_17437205 | 3.74 |
ENSMUST00000006053.13
ENSMUST00000231311.2 ENSMUST00000163476.10 ENSMUST00000231257.2 ENSMUST00000165363.10 ENSMUST00000232043.2 ENSMUST00000090159.13 ENSMUST00000232442.2 |
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr6_+_42326760 | 3.73 |
ENSMUST00000203652.3
ENSMUST00000070635.13 |
Zyx
|
zyxin |
| chr10_+_127159568 | 3.71 |
ENSMUST00000219026.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr7_+_65710086 | 3.68 |
ENSMUST00000153609.8
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr13_-_38842967 | 3.67 |
ENSMUST00000001757.9
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr3_-_54823287 | 3.67 |
ENSMUST00000070342.4
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr7_-_6699422 | 3.66 |
ENSMUST00000122432.4
ENSMUST00000002336.16 |
Zim1
|
zinc finger, imprinted 1 |
| chr16_-_4536992 | 3.65 |
ENSMUST00000115851.10
|
Nmral1
|
NmrA-like family domain containing 1 |
| chr5_+_114924106 | 3.61 |
ENSMUST00000137519.2
|
Ankrd13a
|
ankyrin repeat domain 13a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 36.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 8.3 | 25.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 6.9 | 20.7 | GO:0070947 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) neutrophil mediated killing of fungus(GO:0070947) |
| 5.8 | 17.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 5.4 | 16.2 | GO:0071846 | actin filament debranching(GO:0071846) |
| 5.2 | 15.6 | GO:0015825 | L-serine transport(GO:0015825) |
| 4.7 | 28.0 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 4.0 | 12.0 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 3.9 | 23.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 3.7 | 18.7 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 3.4 | 13.8 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 3.4 | 10.2 | GO:0030221 | basophil differentiation(GO:0030221) |
| 3.0 | 12.0 | GO:0036233 | glycine import(GO:0036233) |
| 2.9 | 11.6 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) positive regulation of osteoclast proliferation(GO:0090290) |
| 2.6 | 15.6 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 2.4 | 19.4 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 2.2 | 6.5 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 2.2 | 6.5 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 2.2 | 10.8 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 2.1 | 6.3 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 2.1 | 22.9 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 2.0 | 7.9 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 1.9 | 7.7 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 1.9 | 5.7 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 1.9 | 7.6 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 1.7 | 5.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.7 | 11.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 1.6 | 4.9 | GO:0032976 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 1.6 | 4.8 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.6 | 27.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.6 | 8.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 1.6 | 4.7 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 1.5 | 18.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.5 | 4.4 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.4 | 5.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.4 | 4.3 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 1.4 | 4.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.3 | 10.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.3 | 4.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 1.3 | 6.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.3 | 23.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 1.3 | 9.1 | GO:0015886 | heme transport(GO:0015886) |
| 1.3 | 3.8 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 1.3 | 3.8 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 1.3 | 3.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.2 | 6.1 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 1.2 | 10.7 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.2 | 8.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 1.1 | 3.4 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 1.1 | 4.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 1.1 | 5.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.1 | 5.5 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 1.1 | 3.3 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.1 | 6.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 1.1 | 4.3 | GO:0017126 | nucleologenesis(GO:0017126) |
| 1.1 | 9.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.0 | 2.0 | GO:0002358 | B cell homeostatic proliferation(GO:0002358) |
| 1.0 | 4.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 1.0 | 2.9 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.0 | 2.9 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.0 | 7.6 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.9 | 2.8 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.9 | 9.4 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.9 | 2.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.9 | 0.9 | GO:0071029 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) |
| 0.9 | 4.4 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.9 | 4.4 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.9 | 6.9 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.9 | 6.0 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.9 | 3.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.9 | 3.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.9 | 2.6 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.8 | 8.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.8 | 3.4 | GO:2001188 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.8 | 2.5 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.8 | 5.8 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.8 | 24.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.8 | 12.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.8 | 2.5 | GO:0046101 | GMP catabolic process(GO:0046038) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.8 | 16.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.8 | 4.7 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.8 | 6.3 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.8 | 3.9 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.8 | 10.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.8 | 2.3 | GO:1990764 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of actin filament-based movement(GO:1903116) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.7 | 5.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.7 | 5.0 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.7 | 2.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.7 | 5.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.7 | 2.8 | GO:0035546 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.7 | 2.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.7 | 13.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.7 | 4.0 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.6 | 13.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.6 | 1.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.6 | 3.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.6 | 5.5 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.6 | 2.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.6 | 3.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.6 | 2.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.6 | 5.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.6 | 3.0 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.6 | 1.7 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.6 | 3.4 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.6 | 2.2 | GO:0015744 | succinate transport(GO:0015744) |
| 0.6 | 12.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.6 | 2.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.6 | 1.7 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.6 | 1.7 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.6 | 1.7 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.5 | 4.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.5 | 4.9 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.5 | 15.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.5 | 1.6 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.5 | 5.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.5 | 3.7 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.5 | 6.8 | GO:1904667 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.5 | 2.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.5 | 5.7 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.5 | 0.5 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.5 | 4.1 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.5 | 7.1 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.5 | 2.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.5 | 1.4 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.5 | 2.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 2.4 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.5 | 2.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.5 | 4.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.4 | 1.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.4 | 10.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.4 | 2.6 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.4 | 1.7 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.4 | 3.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.4 | 2.0 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.4 | 1.6 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.4 | 8.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.4 | 1.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.4 | 5.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.4 | 1.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.4 | 17.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.4 | 3.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 1.1 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.4 | 7.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.4 | 3.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.4 | 0.7 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.4 | 1.8 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.4 | 1.8 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.4 | 0.7 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.4 | 2.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 15.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.3 | 2.8 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 3.8 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.3 | 0.3 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.3 | 1.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.3 | 1.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 1.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.3 | 3.4 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.3 | 11.2 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.3 | 13.4 | GO:0055069 | zinc ion homeostasis(GO:0055069) |
| 0.3 | 1.0 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.3 | 1.0 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) negative regulation of interleukin-18 production(GO:0032701) |
| 0.3 | 1.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.3 | 3.5 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.3 | 3.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 3.2 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 1.3 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.3 | 5.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 1.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.3 | 4.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 1.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.3 | 1.5 | GO:1903936 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) cellular response to sodium arsenite(GO:1903936) |
| 0.3 | 3.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 4.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.3 | 1.2 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 | 0.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.3 | 0.9 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.3 | 1.4 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.3 | 0.9 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.3 | 7.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 3.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 2.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 | 1.7 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 0.3 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.3 | 13.0 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.3 | 5.4 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.3 | 2.4 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.3 | 0.8 | GO:2000642 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 2.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.3 | 0.5 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.3 | 2.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.3 | 2.3 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.3 | 2.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 2.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 1.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 1.7 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 1.7 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 | 1.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 2.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 8.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 8.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 1.8 | GO:0033277 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 0.9 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 2.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.7 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 5.2 | GO:0019081 | viral translation(GO:0019081) |
| 0.2 | 0.9 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.2 | 1.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 1.5 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 10.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 0.8 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.2 | 5.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 2.5 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.2 | 9.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 2.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 1.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.2 | 2.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.2 | 4.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 2.4 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.2 | 2.9 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.2 | 1.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 1.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 2.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 1.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 2.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 4.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 4.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 7.9 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.2 | 0.4 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.2 | 7.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 1.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 0.9 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 0.4 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.2 | 1.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.5 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 0.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 10.9 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 2.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 1.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 0.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 | 1.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 3.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.2 | 0.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 3.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 2.5 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 3.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.5 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.2 | 0.9 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.2 | 2.6 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.2 | 3.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.2 | 0.8 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.2 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 9.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 1.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 24.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.9 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 2.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 2.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 3.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.7 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 3.9 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.6 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 1.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 3.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 6.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 5.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 2.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 8.0 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 | 0.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 5.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 3.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 0.8 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.1 | 0.3 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 2.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 2.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 3.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 0.3 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 2.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 5.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 1.4 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 2.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 4.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 2.0 | GO:0031297 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.1 | 1.8 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.4 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.9 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 1.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.4 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 1.2 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 3.9 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.9 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 1.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 12.9 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 1.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 2.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 17.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 3.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 4.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.3 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.3 | GO:0035565 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.6 | GO:0061370 | estrogen biosynthetic process(GO:0006703) testosterone biosynthetic process(GO:0061370) |
| 0.1 | 1.7 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.1 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 2.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 1.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 3.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.2 | GO:1900738 | psychomotor behavior(GO:0036343) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.5 | GO:0090042 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.7 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 0.2 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 | 0.3 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.1 | 0.4 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.9 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 2.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.0 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.3 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 1.3 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 1.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 3.3 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 1.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 2.6 | GO:0007088 | regulation of mitotic nuclear division(GO:0007088) |
| 0.0 | 0.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 2.5 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 1.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.7 | GO:0002076 | osteoblast development(GO:0002076) negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 1.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.6 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0070829 | chromatin maintenance(GO:0070827) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.3 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 1.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 1.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.2 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.8 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.6 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 1.0 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.6 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.4 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.2 | GO:0051642 | centrosome localization(GO:0051642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 28.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 2.7 | 10.9 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 2.2 | 15.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 2.2 | 6.6 | GO:0070992 | translation initiation complex(GO:0070992) |
| 1.7 | 8.7 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.7 | 8.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 1.7 | 25.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.6 | 18.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 1.6 | 4.9 | GO:0097144 | BAX complex(GO:0097144) |
| 1.6 | 4.7 | GO:1990879 | CST complex(GO:1990879) |
| 1.5 | 4.6 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 1.5 | 5.9 | GO:0032021 | NELF complex(GO:0032021) |
| 1.4 | 5.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.3 | 6.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.3 | 29.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 1.2 | 8.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.1 | 3.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 1.1 | 18.0 | GO:0044754 | autolysosome(GO:0044754) |
| 1.1 | 5.5 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 1.1 | 10.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.0 | 12.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.0 | 5.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.0 | 2.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.8 | 51.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.8 | 3.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.8 | 2.3 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.7 | 7.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.7 | 4.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.7 | 14.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.7 | 5.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.7 | 4.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.7 | 10.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.6 | 3.9 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.6 | 3.8 | GO:1990393 | 3M complex(GO:1990393) |
| 0.6 | 14.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.6 | 10.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.6 | 44.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 2.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.6 | 6.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.6 | 18.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.6 | 1.7 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.5 | 3.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 9.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.5 | 17.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.5 | 7.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 11.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 7.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 2.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.5 | 6.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.5 | 9.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.5 | 1.5 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.5 | 1.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.5 | 1.9 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.5 | 18.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.5 | 1.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.5 | 2.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.5 | 3.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.5 | 2.8 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.5 | 1.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.5 | 4.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.4 | 6.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 10.0 | GO:0097526 | spliceosomal tri-snRNP complex(GO:0097526) |
| 0.4 | 2.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.4 | 7.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.4 | 2.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.4 | 1.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.4 | 2.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 11.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.4 | 5.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.4 | 3.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.4 | 1.1 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.4 | 3.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 2.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 1.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.4 | 6.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.4 | 15.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.4 | 4.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.4 | 2.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.3 | 1.7 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.3 | 2.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.3 | 2.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.3 | 0.3 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.3 | 2.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.3 | 4.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 3.7 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 4.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 1.5 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.3 | 1.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 7.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 1.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 11.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 2.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.3 | 1.7 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 1.4 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.3 | 1.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 3.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 5.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 1.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 1.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 3.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 2.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 4.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 1.9 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.2 | 2.8 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 8.1 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 1.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 6.1 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 3.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 3.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 3.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 14.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 27.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 2.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 10.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 0.6 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 3.5 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.2 | 0.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 1.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 5.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 24.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 0.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.2 | 1.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 15.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.2 | 3.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 3.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.9 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 4.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 3.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 13.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 1.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 4.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 1.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 1.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 4.0 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 3.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.8 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 3.3 | GO:0030684 | preribosome(GO:0030684) |
| 0.1 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 27.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 2.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 4.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 4.5 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 4.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 4.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 5.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 2.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.9 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 2.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 11.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 2.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 13.6 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 5.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0034719 | SMN complex(GO:0032797) SMN-Sm protein complex(GO:0034719) |
| 0.0 | 2.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 5.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 20.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 29.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 10.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.6 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 6.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 27.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 2.9 | 8.7 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 2.8 | 2.8 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 2.7 | 8.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 2.2 | 17.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 2.1 | 12.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 2.0 | 8.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 1.9 | 7.7 | GO:0032093 | SAM domain binding(GO:0032093) |
| 1.8 | 14.4 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.8 | 12.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.6 | 14.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.5 | 4.6 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 1.5 | 9.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.4 | 12.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.3 | 4.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.3 | 6.5 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 1.3 | 3.8 | GO:0090556 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 1.3 | 23.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.3 | 7.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.2 | 13.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 1.2 | 7.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 1.2 | 15.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 1.2 | 16.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.2 | 4.7 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.2 | 5.9 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 1.1 | 11.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.1 | 3.4 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.1 | 3.3 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 1.1 | 6.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.0 | 3.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.0 | 18.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.0 | 23.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 1.0 | 10.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.0 | 3.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.9 | 4.6 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.9 | 2.8 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.9 | 10.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.9 | 11.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.9 | 6.0 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.8 | 3.3 | GO:0072354 | histone kinase activity (H3-T3 specific)(GO:0072354) |
| 0.8 | 4.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.8 | 2.4 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.8 | 10.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.7 | 2.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.7 | 2.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.7 | 2.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.7 | 2.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.6 | 6.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.6 | 4.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 5.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.6 | 0.6 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.6 | 7.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.6 | 7.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.6 | 25.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.6 | 5.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.6 | 4.0 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.6 | 5.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.6 | 7.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.6 | 1.7 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 1.6 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.5 | 3.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.5 | 3.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.5 | 4.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 4.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.5 | 4.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.5 | 1.5 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.5 | 2.4 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.5 | 4.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 2.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.5 | 2.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.5 | 8.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.5 | 5.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 82.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.4 | 3.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.4 | 1.7 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.4 | 10.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 1.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.4 | 2.8 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.4 | 18.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 3.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 20.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.4 | 1.6 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.4 | 15.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.4 | 1.5 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.4 | 3.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.4 | 3.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 1.8 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.4 | 6.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.4 | 1.4 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.4 | 35.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 3.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.3 | 1.7 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.3 | 2.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 12.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.3 | 2.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 2.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 1.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 4.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.3 | 4.7 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.3 | 1.9 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.3 | 1.5 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.3 | 4.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.3 | 1.8 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.3 | 8.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 1.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 1.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.3 | 0.9 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.3 | 4.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.3 | 4.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 2.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 6.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 1.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 2.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 1.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 1.9 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.3 | 8.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.3 | 6.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 13.6 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.3 | 0.8 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.3 | 1.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 3.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 6.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 8.6 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.2 | 2.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 3.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 16.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.2 | 2.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.2 | 5.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 4.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 6.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 5.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 2.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 4.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 0.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 5.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 10.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 1.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 14.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 2.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 5.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 5.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 4.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 2.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 2.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 3.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 2.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 1.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 2.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 2.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 4.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 2.0 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.2 | 1.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 3.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.7 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 6.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 5.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 2.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.8 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 5.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 11.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 6.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 2.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 2.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 20.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 2.9 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 3.7 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 2.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 4.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 14.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.4 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.3 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 1.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.7 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 5.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 4.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 0.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 2.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 4.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.6 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 2.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.0 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 6.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.8 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 10.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 4.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 2.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 2.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.8 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.1 | 3.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 6.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 3.6 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 3.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 4.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 3.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 8.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 1.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 2.2 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 22.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 5.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 17.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 1.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 2.0 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 5.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.4 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 45.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 1.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.5 | 25.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 18.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 24.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.4 | 14.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 20.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 31.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 18.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 13.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 5.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 8.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 12.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 11.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 2.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 11.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 21.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 5.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 7.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 5.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 14.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 21.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 9.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 4.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 1.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 10.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 4.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 7.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 4.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 5.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 9.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 6.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 3.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.7 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 5.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 4.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.2 | 23.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 1.1 | 18.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.0 | 25.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.8 | 104.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.8 | 24.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.7 | 21.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.7 | 45.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.6 | 33.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.6 | 10.0 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.6 | 1.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 11.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.5 | 2.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 15.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 13.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 12.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 13.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.4 | 5.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.4 | 6.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 7.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 11.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.4 | 9.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 33.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 18.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 4.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.3 | 20.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.3 | 2.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 5.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 5.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 4.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 7.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 5.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.3 | 5.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.3 | 4.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.2 | 4.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 4.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 5.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 26.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 12.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 2.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 11.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 6.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 1.6 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.2 | 4.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 3.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 20.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 5.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 6.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 5.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 8.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 5.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 4.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 5.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 5.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 11.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 3.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 5.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 11.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.0 | 0.7 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 2.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 4.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |