Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
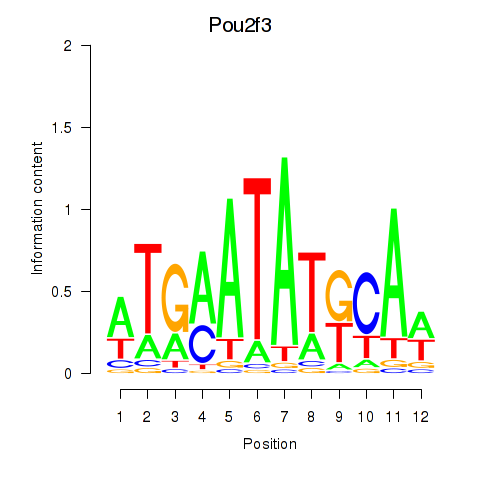
Results for Pou2f3
Z-value: 1.53
Transcription factors associated with Pou2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f3
|
ENSMUSG00000032015.17 | Pou2f3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f3 | mm39_v1_chr9_-_43117052_43117148 | 0.31 | 7.0e-02 | Click! |
Activity profile of Pou2f3 motif
Sorted Z-values of Pou2f3 motif
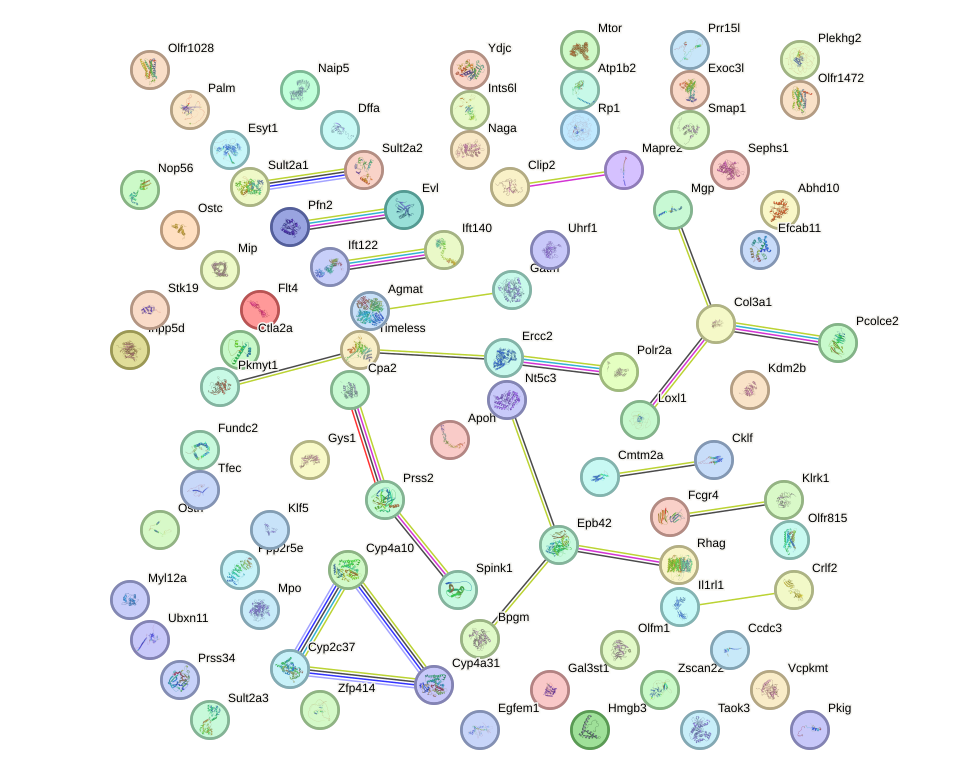
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13467422 | 9.96 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr7_-_13571334 | 9.73 |
ENSMUST00000108522.5
|
Sult2a1
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr4_+_115420876 | 5.13 |
ENSMUST00000126645.8
ENSMUST00000030480.4 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr4_+_115420817 | 4.78 |
ENSMUST00000141033.8
ENSMUST00000030486.15 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr11_+_87684299 | 3.98 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr6_+_41498716 | 3.61 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr1_+_87603952 | 3.55 |
ENSMUST00000170300.8
ENSMUST00000167032.2 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr6_+_30541581 | 3.40 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr7_-_142209755 | 2.85 |
ENSMUST00000178921.2
|
Igf2
|
insulin-like growth factor 2 |
| chr7_+_12631727 | 2.70 |
ENSMUST00000055528.11
ENSMUST00000117189.2 ENSMUST00000120809.2 ENSMUST00000119989.3 |
Zscan22
|
zinc finger and SCAN domain containing 22 |
| chr16_-_18904240 | 2.69 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr17_+_41121979 | 2.47 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr11_-_69496655 | 2.21 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr6_-_68713748 | 2.09 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr17_+_25517363 | 2.07 |
ENSMUST00000037453.4
|
Prss34
|
protease, serine 34 |
| chr19_+_39980868 | 2.07 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr12_+_108572015 | 2.05 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr12_-_114355789 | 1.93 |
ENSMUST00000103486.2
|
Ighv6-3
|
immunoglobulin heavy variable 6-3 |
| chr6_+_34453142 | 1.85 |
ENSMUST00000045372.6
ENSMUST00000138668.2 ENSMUST00000139067.2 |
Bpgm
|
2,3-bisphosphoglycerate mutase |
| chr11_+_3939924 | 1.83 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr9_-_58220469 | 1.75 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr11_-_69649452 | 1.72 |
ENSMUST00000058470.16
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr18_-_43870622 | 1.71 |
ENSMUST00000025381.4
|
Spink1
|
serine peptidase inhibitor, Kazal type 1 |
| chr7_-_126014027 | 1.67 |
ENSMUST00000032968.7
ENSMUST00000206325.2 |
Cd19
|
CD19 antigen |
| chr1_+_45350698 | 1.57 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr6_-_69800923 | 1.51 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr17_+_25235310 | 1.47 |
ENSMUST00000024983.12
|
Ift140
|
intraflagellar transport 140 |
| chr19_+_40648182 | 1.46 |
ENSMUST00000112231.9
ENSMUST00000127828.8 |
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chrX_-_23132991 | 1.44 |
ENSMUST00000115316.9
|
Klhl13
|
kelch-like 13 |
| chr7_-_13856967 | 1.43 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr12_-_114752425 | 1.42 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr3_+_142406787 | 1.37 |
ENSMUST00000106218.8
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr2_+_5142567 | 1.36 |
ENSMUST00000027988.8
|
Ccdc3
|
coiled-coil domain containing 3 |
| chr12_-_114621406 | 1.35 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr3_+_142406827 | 1.35 |
ENSMUST00000044392.11
ENSMUST00000199519.5 |
Kyat3
|
kynurenine aminotransferase 3 |
| chr17_-_71305003 | 1.32 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr16_+_16964801 | 1.30 |
ENSMUST00000232479.2
ENSMUST00000232344.2 ENSMUST00000069064.7 |
Ydjc
|
YdjC homolog (bacterial) |
| chrX_+_55500170 | 1.29 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr7_-_28071919 | 1.29 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr10_-_75776437 | 1.26 |
ENSMUST00000219979.2
|
Gm867
|
predicted gene 867 |
| chr17_+_33848054 | 1.25 |
ENSMUST00000166627.8
ENSMUST00000073570.12 ENSMUST00000170225.3 |
Zfp414
|
zinc finger protein 414 |
| chr13_-_61084358 | 1.20 |
ENSMUST00000225859.2
ENSMUST00000225167.2 ENSMUST00000021880.10 |
Gm49391
Ctla2a
|
predicted gene, 49391 cytotoxic T lymphocyte-associated protein 2 alpha |
| chr12_-_69629758 | 1.17 |
ENSMUST00000058639.11
|
Vcpkmt
|
valosin containing protein lysine (K) methyltransferase |
| chr3_+_20039775 | 1.17 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr10_+_79650496 | 1.13 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr2_+_163535925 | 1.12 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr5_-_109706801 | 1.12 |
ENSMUST00000200284.5
ENSMUST00000044579.12 |
Crlf2
|
cytokine receptor-like factor 2 |
| chr12_-_114252202 | 1.11 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr17_+_23945310 | 1.10 |
ENSMUST00000024701.9
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr12_-_113542610 | 1.08 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr1_+_170846482 | 1.08 |
ENSMUST00000078825.5
|
Fcgr4
|
Fc receptor, IgG, low affinity IV |
| chr6_-_16898440 | 1.07 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr2_-_120867232 | 1.05 |
ENSMUST00000023987.6
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr17_+_25235039 | 1.05 |
ENSMUST00000142000.9
ENSMUST00000137386.8 |
Ift140
|
intraflagellar transport 140 |
| chr11_+_96820091 | 1.03 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr12_-_114443071 | 0.99 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr2_+_130119077 | 0.97 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chrX_+_70599524 | 0.96 |
ENSMUST00000072699.13
ENSMUST00000114582.9 ENSMUST00000015361.11 ENSMUST00000088874.10 |
Hmgb3
|
high mobility group box 3 |
| chr12_-_113958518 | 0.95 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr12_-_114140482 | 0.95 |
ENSMUST00000103475.2
ENSMUST00000195706.2 |
Ighv14-4
|
immunoglobulin heavy variable 14-4 |
| chr9_+_95519654 | 0.94 |
ENSMUST00000015498.9
|
Pcolce2
|
procollagen C-endopeptidase enhancer 2 |
| chr11_+_108286114 | 0.91 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr10_+_128067964 | 0.91 |
ENSMUST00000125289.8
ENSMUST00000105242.8 |
Timeless
|
timeless circadian clock 1 |
| chr17_+_56610321 | 0.91 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr12_-_115172211 | 0.90 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr4_+_141473983 | 0.90 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr7_-_28072022 | 0.89 |
ENSMUST00000144700.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr6_+_115830431 | 0.88 |
ENSMUST00000112925.8
ENSMUST00000038234.13 ENSMUST00000112923.7 |
Ift122
|
intraflagellar transport 122 |
| chr11_+_96820220 | 0.88 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr7_+_45084257 | 0.86 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr12_-_75596441 | 0.83 |
ENSMUST00000218716.2
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr7_-_28071658 | 0.83 |
ENSMUST00000094644.11
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr17_+_56610396 | 0.82 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr6_-_136852792 | 0.82 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr8_-_106022676 | 0.81 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chr11_+_49500090 | 0.81 |
ENSMUST00000020617.3
|
Flt4
|
FMS-like tyrosine kinase 4 |
| chr7_+_108265625 | 0.80 |
ENSMUST00000213979.3
ENSMUST00000216331.2 ENSMUST00000217170.2 |
Olfr510
|
olfactory receptor 510 |
| chr4_+_149188585 | 0.79 |
ENSMUST00000103216.10
ENSMUST00000030816.4 |
Dffa
|
DNA fragmentation factor, alpha subunit |
| chr2_-_152672535 | 0.79 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr12_-_114547622 | 0.76 |
ENSMUST00000193893.6
ENSMUST00000103498.3 |
Ighv1-9
|
immunoglobulin heavy variable V1-9 |
| chr6_-_56878854 | 0.75 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr10_-_128361731 | 0.75 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr12_-_75678092 | 0.74 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr2_-_122441719 | 0.74 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr1_-_23948764 | 0.74 |
ENSMUST00000129254.8
|
Smap1
|
small ArfGAP 1 |
| chrX_+_74425990 | 0.72 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr10_-_78131228 | 0.71 |
ENSMUST00000105387.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr10_-_129738595 | 0.69 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr12_-_114023935 | 0.69 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr5_+_117378510 | 0.68 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr1_+_39940189 | 0.67 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr2_+_85780781 | 0.67 |
ENSMUST00000080698.3
|
Olfr1028
|
olfactory receptor 1028 |
| chr16_-_45563250 | 0.65 |
ENSMUST00000066983.13
|
Abhd10
|
abhydrolase domain containing 10 |
| chr12_-_115722081 | 0.65 |
ENSMUST00000103541.3
|
Ighv1-72
|
immunoglobulin heavy variable 1-72 |
| chr1_-_171476495 | 0.65 |
ENSMUST00000194791.2
ENSMUST00000192024.6 |
Slamf7
|
SLAM family member 7 |
| chr5_-_123038329 | 0.65 |
ENSMUST00000031435.14
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr12_-_115122455 | 0.64 |
ENSMUST00000103523.2
|
Ighv1-53
|
immunoglobulin heavy variable 1-53 |
| chr4_+_148533062 | 0.63 |
ENSMUST00000103221.10
ENSMUST00000057580.8 |
Mtor
|
mechanistic target of rapamycin kinase |
| chr1_-_4479445 | 0.62 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr12_-_114502585 | 0.62 |
ENSMUST00000103496.4
|
Ighv1-7
|
immunoglobulin heavy variable V1-7 |
| chr12_-_115471634 | 0.61 |
ENSMUST00000103535.3
|
Ighv1-64
|
immunoglobulin heavy variable 1-64 |
| chr12_-_114901026 | 0.61 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr7_-_45084012 | 0.60 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr1_+_40478787 | 0.60 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr5_-_134581235 | 0.59 |
ENSMUST00000036999.10
ENSMUST00000100647.7 |
Clip2
|
CAP-GLY domain containing linker protein 2 |
| chr15_-_82223065 | 0.59 |
ENSMUST00000229733.2
ENSMUST00000229388.2 |
Naga
|
N-acetyl galactosaminidase, alpha |
| chr2_+_86122799 | 0.58 |
ENSMUST00000217166.2
|
Olfr1052
|
olfactory receptor 1052 |
| chr2_+_4887015 | 0.58 |
ENSMUST00000115019.2
|
Sephs1
|
selenophosphate synthetase 1 |
| chr1_+_39940043 | 0.57 |
ENSMUST00000168431.7
ENSMUST00000163854.9 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr2_-_152672185 | 0.57 |
ENSMUST00000140436.2
|
Bcl2l1
|
BCL2-like 1 |
| chr3_-_130503041 | 0.55 |
ENSMUST00000043937.9
|
Ostc
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr13_-_100382831 | 0.55 |
ENSMUST00000049789.3
|
Naip5
|
NLR family, apoptosis inhibitory protein 5 |
| chr12_-_115157739 | 0.53 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr2_+_28083105 | 0.53 |
ENSMUST00000100244.10
|
Olfm1
|
olfactomedin 1 |
| chr12_-_99849660 | 0.52 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr12_-_114646685 | 0.51 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr7_-_45083688 | 0.51 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr6_-_69704122 | 0.51 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr19_+_13478481 | 0.50 |
ENSMUST00000214274.2
|
Olfr1477
|
olfactory receptor 1477 |
| chr12_-_114477427 | 0.50 |
ENSMUST00000191803.2
|
Ighv1-5
|
immunoglobulin heavy variable V1-5 |
| chr7_+_19115929 | 0.49 |
ENSMUST00000062831.16
ENSMUST00000108461.8 ENSMUST00000108460.8 |
Ercc2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr3_-_57755500 | 0.49 |
ENSMUST00000066882.10
|
Pfn2
|
profilin 2 |
| chr4_+_133829898 | 0.49 |
ENSMUST00000070246.9
ENSMUST00000156750.2 |
Ubxn11
|
UBX domain protein 11 |
| chr12_-_115884332 | 0.48 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr12_-_113896002 | 0.47 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr6_-_129599645 | 0.47 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr8_+_104977493 | 0.47 |
ENSMUST00000034342.13
ENSMUST00000212433.2 ENSMUST00000211809.2 |
Cklf
|
chemokine-like factor |
| chr1_-_171476559 | 0.47 |
ENSMUST00000111276.9
ENSMUST00000194531.6 |
Slamf7
|
SLAM family member 7 |
| chrX_-_36255377 | 0.46 |
ENSMUST00000152291.3
|
Septin6
|
septin 6 |
| chr6_-_69753317 | 0.46 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr12_+_98234884 | 0.46 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr2_-_110193502 | 0.46 |
ENSMUST00000099626.5
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr7_+_126895463 | 0.46 |
ENSMUST00000106306.9
ENSMUST00000120857.8 |
Itgal
|
integrin alpha L |
| chr16_+_18317463 | 0.46 |
ENSMUST00000231621.2
ENSMUST00000139625.8 |
Gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr1_+_171668173 | 0.46 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr9_-_21199232 | 0.46 |
ENSMUST00000184326.8
ENSMUST00000038671.10 |
Kri1
|
KRI1 homolog |
| chr7_+_65710086 | 0.45 |
ENSMUST00000153609.8
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr2_+_163916042 | 0.45 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr2_+_28082943 | 0.45 |
ENSMUST00000113920.8
|
Olfm1
|
olfactomedin 1 |
| chr12_-_54842488 | 0.45 |
ENSMUST00000005798.9
|
Snx6
|
sorting nexin 6 |
| chr4_+_152123772 | 0.45 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr4_-_117354249 | 0.44 |
ENSMUST00000030439.15
|
Rnf220
|
ring finger protein 220 |
| chr12_-_115587215 | 0.44 |
ENSMUST00000199933.5
ENSMUST00000103539.3 |
Ighv1-69
|
immunoglobulin heavy variable 1-69 |
| chr12_-_114263874 | 0.43 |
ENSMUST00000103482.2
ENSMUST00000194159.2 |
Ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr6_-_69415741 | 0.43 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr11_-_60770098 | 0.43 |
ENSMUST00000062677.12
|
Tmem11
|
transmembrane protein 11 |
| chr6_+_68279392 | 0.43 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr12_+_30961650 | 0.42 |
ENSMUST00000020997.15
ENSMUST00000110880.3 |
Sh3yl1
|
Sh3 domain YSC-like 1 |
| chr7_+_126895423 | 0.42 |
ENSMUST00000117762.8
|
Itgal
|
integrin alpha L |
| chr18_+_37853415 | 0.42 |
ENSMUST00000195363.2
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr9_-_19799300 | 0.41 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chr9_-_45923908 | 0.41 |
ENSMUST00000217514.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr1_+_40478926 | 0.41 |
ENSMUST00000173514.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr12_-_114710326 | 0.40 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr12_-_115299134 | 0.39 |
ENSMUST00000195359.6
ENSMUST00000103530.3 |
Ighv1-59
|
immunoglobulin heavy variable V1-59 |
| chr19_-_36896999 | 0.39 |
ENSMUST00000238948.2
ENSMUST00000057337.9 |
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr4_+_102446883 | 0.39 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr4_-_129517662 | 0.38 |
ENSMUST00000106035.8
ENSMUST00000150357.2 ENSMUST00000030586.15 |
Ccdc28b
|
coiled coil domain containing 28B |
| chr1_-_79417732 | 0.38 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr7_+_44146029 | 0.37 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr6_-_48685108 | 0.37 |
ENSMUST00000126422.3
ENSMUST00000119315.2 ENSMUST00000053661.7 |
Gimap6
|
GTPase, IMAP family member 6 |
| chr10_-_126866658 | 0.37 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr2_+_85838122 | 0.36 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
| chr2_-_86528739 | 0.36 |
ENSMUST00000214141.2
|
Olfr1087
|
olfactory receptor 1087 |
| chr8_-_86091946 | 0.35 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr16_-_59092995 | 0.35 |
ENSMUST00000216834.2
|
Olfr201
|
olfactory receptor 201 |
| chr11_+_98632631 | 0.35 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr9_+_5308828 | 0.34 |
ENSMUST00000162846.8
ENSMUST00000027012.14 |
Casp4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr10_+_89906956 | 0.34 |
ENSMUST00000183109.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_+_20524565 | 0.34 |
ENSMUST00000089217.11
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr7_+_103197281 | 0.34 |
ENSMUST00000214173.2
|
Olfr613
|
olfactory receptor 613 |
| chr6_+_68657317 | 0.34 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr9_+_121946404 | 0.33 |
ENSMUST00000134949.8
|
Snrk
|
SNF related kinase |
| chr6_-_130208601 | 0.33 |
ENSMUST00000088011.11
ENSMUST00000112013.8 ENSMUST00000049304.14 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr3_+_87283687 | 0.33 |
ENSMUST00000163661.8
ENSMUST00000072480.9 |
Fcrl1
|
Fc receptor-like 1 |
| chr17_+_80614795 | 0.33 |
ENSMUST00000223878.2
ENSMUST00000068175.6 ENSMUST00000224391.2 |
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr8_-_34574970 | 0.33 |
ENSMUST00000118811.8
|
Dctn6
|
dynactin 6 |
| chr10_-_126866682 | 0.32 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr2_-_85593110 | 0.32 |
ENSMUST00000214958.2
|
Olfr1012
|
olfactory receptor 1012 |
| chr14_-_50479161 | 0.32 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr6_-_101176147 | 0.31 |
ENSMUST00000239140.2
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr11_+_22462088 | 0.31 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr6_+_125192514 | 0.31 |
ENSMUST00000032487.14
ENSMUST00000100942.9 ENSMUST00000063588.11 |
Vamp1
|
vesicle-associated membrane protein 1 |
| chrX_+_149377416 | 0.31 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_-_27765381 | 0.30 |
ENSMUST00000193779.2
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr12_-_115766700 | 0.30 |
ENSMUST00000196587.5
ENSMUST00000103543.3 |
Ighv1-74
|
immunoglobulin heavy variable V1-74 |
| chr12_-_115109539 | 0.30 |
ENSMUST00000192554.6
ENSMUST00000103522.3 |
Ighv1-52
|
immunoglobulin heavy variable 1-52 |
| chr18_+_37801971 | 0.30 |
ENSMUST00000193869.2
|
Pcdhga2
|
protocadherin gamma subfamily A, 2 |
| chr7_-_107856399 | 0.29 |
ENSMUST00000211508.3
|
Olfr488
|
olfactory receptor 488 |
| chr7_+_27869115 | 0.29 |
ENSMUST00000042405.8
|
Fbl
|
fibrillarin |
| chr5_+_31609772 | 0.28 |
ENSMUST00000202061.4
ENSMUST00000076264.8 ENSMUST00000201450.4 |
Zfp512
|
zinc finger protein 512 |
| chr4_-_45108038 | 0.28 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr7_+_126895531 | 0.28 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr17_-_37523969 | 0.28 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr9_+_36743980 | 0.28 |
ENSMUST00000034630.15
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr6_-_115830307 | 0.27 |
ENSMUST00000032469.13
|
Mbd4
|
methyl-CpG binding domain protein 4 |
| chrX_+_162923474 | 0.27 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr12_-_115083839 | 0.27 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr1_+_87331427 | 0.27 |
ENSMUST00000172736.8
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr13_+_49658249 | 0.27 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.9 | 3.5 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.6 | 2.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.5 | 2.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.4 | 1.7 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.4 | 9.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.4 | 1.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.3 | 1.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.3 | 1.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 0.9 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 0.7 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.2 | 1.5 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 3.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 2.8 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.9 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 0.7 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 0.6 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.2 | 1.8 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.2 | 1.1 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 | 0.9 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 0.7 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 | 0.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 2.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 2.7 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.2 | 2.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 0.6 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 1.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 3.5 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.2 | 0.5 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.1 | 0.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 1.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 18.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.8 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 | 0.9 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 1.7 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 1.4 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 0.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.1 | 1.0 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.3 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.1 | 1.8 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 0.3 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 1.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 1.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.6 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 1.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 1.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 1.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 1.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.8 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 0.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 8.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.0 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 1.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.7 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 1.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.0 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.3 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.4 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 3.2 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.1 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 2.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 4.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 1.2 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.2 | 18.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.9 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 1.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 9.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 5.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 2.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.2 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.2 | GO:0050656 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 1.4 | 10.1 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.6 | 1.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.5 | 2.7 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.5 | 1.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.4 | 3.5 | GO:0051425 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 0.3 | 1.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 0.9 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 1.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 1.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 1.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.2 | 0.6 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.2 | 2.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.6 | GO:0008456 | alpha-N-acetylgalactosaminidase activity(GO:0008456) |
| 0.2 | 0.6 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.2 | 1.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.7 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.2 | 0.5 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 1.5 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 1.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.2 | 0.5 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.2 | 0.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 9.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 18.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 3.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.5 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.2 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 3.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 4.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 5.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 3.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 4.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 1.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.7 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 9.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.9 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 3.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 3.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |