Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
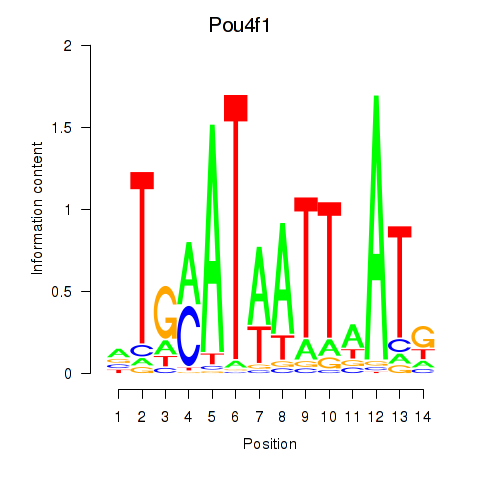
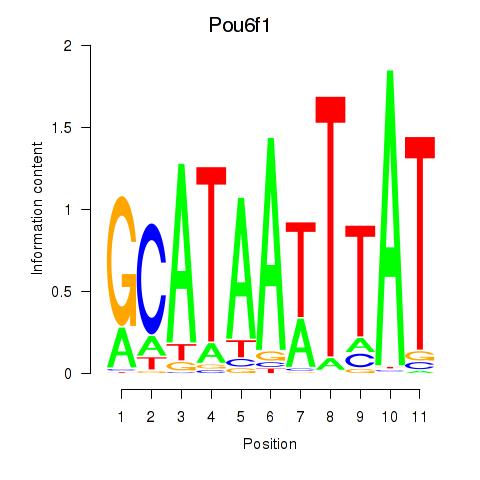
Results for Pou4f1_Pou6f1
Z-value: 0.93
Transcription factors associated with Pou4f1_Pou6f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f1
|
ENSMUSG00000048349.10 | Pou4f1 |
|
Pou6f1
|
ENSMUSG00000009739.18 | Pou6f1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou6f1 | mm39_v1_chr15_-_100497863_100497920 | -0.36 | 3.3e-02 | Click! |
| Pou4f1 | mm39_v1_chr14_-_104705420_104705479 | -0.32 | 5.5e-02 | Click! |
Activity profile of Pou4f1_Pou6f1 motif
Sorted Z-values of Pou4f1_Pou6f1 motif
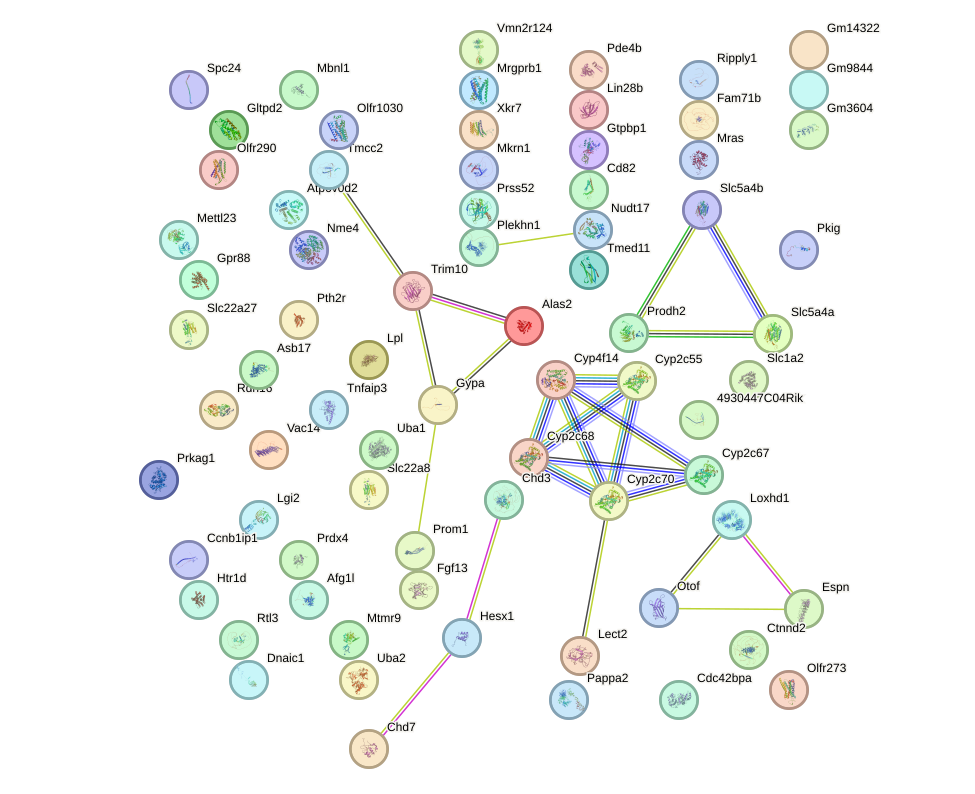
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou4f1_Pou6f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41569775 | 1.69 |
ENSMUST00000102963.10
|
Dnai1
|
dynein axonemal intermediate chain 1 |
| chr5_-_44259010 | 1.14 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr5_-_44259293 | 1.11 |
ENSMUST00000074113.13
|
Prom1
|
prominin 1 |
| chr19_-_7943365 | 0.97 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chrX_-_105884178 | 0.73 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chrX_+_20529137 | 0.73 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr5_-_44259374 | 0.68 |
ENSMUST00000171543.8
|
Prom1
|
prominin 1 |
| chr19_-_40175709 | 0.63 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr19_+_38995463 | 0.61 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr4_-_19922599 | 0.58 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr1_-_179572765 | 0.52 |
ENSMUST00000211943.3
ENSMUST00000131716.4 ENSMUST00000221136.2 |
Kif28
|
kinesin family member 28 |
| chr13_+_23879775 | 0.50 |
ENSMUST00000041052.5
|
H1f6
|
H1.6 linker histone, cluster member |
| chr13_-_56696222 | 0.48 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr15_-_98729333 | 0.46 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr14_-_51045182 | 0.45 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr10_-_75946790 | 0.44 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr15_+_79575046 | 0.44 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr8_+_69333143 | 0.42 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr3_+_153549846 | 0.42 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr17_-_33136277 | 0.41 |
ENSMUST00000234538.2
ENSMUST00000235058.2 ENSMUST00000234759.2 ENSMUST00000179434.8 ENSMUST00000234797.2 |
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr13_-_56696310 | 0.39 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_+_179788037 | 0.38 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr17_-_33136021 | 0.38 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr9_+_56344700 | 0.38 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr6_-_102441628 | 0.38 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr3_-_33136153 | 0.37 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr13_+_19398273 | 0.37 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr3_-_63391300 | 0.36 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr4_+_13784749 | 0.35 |
ENSMUST00000098256.4
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr9_-_99302205 | 0.34 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr14_+_26722319 | 0.34 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr5_-_108943211 | 0.33 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr3_+_60380243 | 0.32 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr10_-_18890281 | 0.31 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr5_-_30619246 | 0.31 |
ENSMUST00000114747.9
ENSMUST00000074171.10 |
Otof
|
otoferlin |
| chr1_+_179788675 | 0.30 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_-_132318039 | 0.30 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr13_-_19491721 | 0.29 |
ENSMUST00000103561.3
|
Trgc2
|
T cell receptor gamma, constant 2 |
| chr7_+_30193047 | 0.29 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chrX_-_138683102 | 0.29 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr2_-_88600832 | 0.29 |
ENSMUST00000217588.3
|
Olfr1200
|
olfactory receptor 1200 |
| chr2_-_93283024 | 0.28 |
ENSMUST00000111257.8
ENSMUST00000145553.8 |
Cd82
|
CD82 antigen |
| chr1_-_80439165 | 0.28 |
ENSMUST00000211023.2
|
Gm45261
|
predicted gene 45261 |
| chr6_-_39397334 | 0.28 |
ENSMUST00000031985.13
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr11_+_116739727 | 0.27 |
ENSMUST00000143184.2
|
Mettl23
|
methyltransferase like 23 |
| chr7_-_100164007 | 0.27 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chrX_-_154123743 | 0.26 |
ENSMUST00000130349.3
|
Prdx4
|
peroxiredoxin 4 |
| chr2_+_163500290 | 0.25 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr2_+_85809620 | 0.24 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr4_-_156319232 | 0.24 |
ENSMUST00000105569.5
|
Klhl17
|
kelch-like 17 |
| chr15_+_30457772 | 0.24 |
ENSMUST00000228282.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chrX_+_149330371 | 0.23 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr13_+_104424359 | 0.23 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr3_-_96615838 | 0.23 |
ENSMUST00000029742.9
ENSMUST00000200387.2 ENSMUST00000171249.6 |
Nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr11_+_70410445 | 0.23 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr7_+_24561616 | 0.23 |
ENSMUST00000170837.3
|
Gm9844
|
predicted pseudogene 9844 |
| chr2_+_97298002 | 0.22 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr5_-_123663440 | 0.22 |
ENSMUST00000197682.6
|
Gm49027
|
predicted gene, 49027 |
| chr17_+_37180437 | 0.22 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr13_-_22403990 | 0.22 |
ENSMUST00000057516.2
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr8_+_94537910 | 0.22 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr2_+_102488985 | 0.21 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr8_+_111345209 | 0.21 |
ENSMUST00000034190.11
|
Vac14
|
Vac14 homolog (S. cerevisiae) |
| chr9_-_21671571 | 0.21 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr7_+_108064533 | 0.21 |
ENSMUST00000217616.3
|
Olfr498
|
olfactory receptor 498 |
| chr8_+_81220410 | 0.21 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr5_+_114427227 | 0.21 |
ENSMUST00000169347.6
|
Myo1h
|
myosin 1H |
| chr4_+_136150835 | 0.20 |
ENSMUST00000088677.5
ENSMUST00000121571.8 ENSMUST00000117699.2 |
Htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D |
| chr3_-_116047148 | 0.20 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr7_-_126391388 | 0.20 |
ENSMUST00000206570.2
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr1_-_158642039 | 0.20 |
ENSMUST00000161589.3
|
Pappa2
|
pappalysin 2 |
| chr10_+_75983285 | 0.20 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr12_+_58258558 | 0.19 |
ENSMUST00000110671.3
ENSMUST00000044299.3 |
Sstr1
|
somatostatin receptor 1 |
| chr17_-_26314438 | 0.19 |
ENSMUST00000236547.2
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr4_+_102112189 | 0.19 |
ENSMUST00000106908.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr18_+_77369654 | 0.19 |
ENSMUST00000096547.11
ENSMUST00000148341.9 ENSMUST00000123410.9 |
Loxhd1
|
lipoxygenase homology domains 1 |
| chr15_+_92059224 | 0.19 |
ENSMUST00000068378.6
|
Cntn1
|
contactin 1 |
| chr10_-_42354276 | 0.19 |
ENSMUST00000151747.8
|
Afg1l
|
AFG1 like ATPase |
| chr19_-_39729431 | 0.18 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr8_+_10056654 | 0.18 |
ENSMUST00000033892.9
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr4_-_52859227 | 0.18 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273 |
| chr13_-_62519750 | 0.18 |
ENSMUST00000107989.7
|
Gm3604
|
predicted gene 3604 |
| chr2_+_67004178 | 0.18 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr10_-_42354482 | 0.18 |
ENSMUST00000041024.15
|
Afg1l
|
AFG1 like ATPase |
| chr8_-_34574970 | 0.18 |
ENSMUST00000118811.8
|
Dctn6
|
dynactin 6 |
| chr12_-_72964646 | 0.18 |
ENSMUST00000044000.12
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr7_-_48105989 | 0.18 |
ENSMUST00000188918.2
|
Mrgprb1
|
MAS-related GPR, member B1 |
| chr2_+_177404691 | 0.18 |
ENSMUST00000119838.9
|
Gm14322
|
predicted gene 14322 |
| chr3_-_37286714 | 0.18 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr5_-_52723607 | 0.17 |
ENSMUST00000199942.5
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr11_+_46295547 | 0.17 |
ENSMUST00000063166.6
|
Fam71b
|
family with sequence similarity 71, member B |
| chr2_+_152873772 | 0.17 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr17_-_26314461 | 0.17 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr10_-_45346297 | 0.17 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr4_+_8690398 | 0.17 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr10_+_127637015 | 0.17 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr4_-_152236664 | 0.17 |
ENSMUST00000030785.15
ENSMUST00000105658.8 ENSMUST00000207676.2 ENSMUST00000105659.9 |
Espn
|
espin |
| chr8_+_10056631 | 0.17 |
ENSMUST00000207792.3
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr7_+_84562283 | 0.17 |
ENSMUST00000216367.2
ENSMUST00000214501.3 |
Olfr290
|
olfactory receptor 290 |
| chr5_-_52723700 | 0.16 |
ENSMUST00000039750.7
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr10_-_129751466 | 0.16 |
ENSMUST00000213438.2
|
Olfr816
|
olfactory receptor 816 |
| chr2_-_86061745 | 0.16 |
ENSMUST00000216056.2
|
Olfr1047
|
olfactory receptor 1047 |
| chr14_+_64341735 | 0.16 |
ENSMUST00000022537.6
|
Prss52
|
protease, serine 52 |
| chr17_+_18269686 | 0.16 |
ENSMUST00000176802.3
|
Vmn2r124
|
vomeronasal 2, receptor 124 |
| chr15_-_77295234 | 0.16 |
ENSMUST00000089452.6
ENSMUST00000081776.11 |
Apol9a
|
apolipoprotein L 9a |
| chr17_-_21216726 | 0.16 |
ENSMUST00000237195.2
ENSMUST00000237629.2 ENSMUST00000056339.3 |
Vmn1r233
|
vomeronasal 1 receptor 233 |
| chr11_-_4544751 | 0.16 |
ENSMUST00000109943.10
|
Mtmr3
|
myotubularin related protein 3 |
| chr16_-_58898368 | 0.16 |
ENSMUST00000216495.3
|
Olfr190
|
olfactory receptor 190 |
| chrX_-_58211440 | 0.16 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr4_+_102111936 | 0.16 |
ENSMUST00000106907.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr1_+_65321215 | 0.16 |
ENSMUST00000140190.8
|
Pth2r
|
parathyroid hormone 2 receptor |
| chr19_+_8568618 | 0.16 |
ENSMUST00000170817.2
ENSMUST00000010251.11 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr5_-_70999547 | 0.16 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr18_+_44403169 | 0.16 |
ENSMUST00000042747.4
|
Npy6r
|
neuropeptide Y receptor Y6 |
| chr11_-_89732091 | 0.16 |
ENSMUST00000238273.3
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr19_-_46661501 | 0.15 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr4_-_156340713 | 0.15 |
ENSMUST00000219393.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr17_+_38112070 | 0.15 |
ENSMUST00000217365.2
|
Olfr124
|
olfactory receptor 124 |
| chr7_-_103428905 | 0.15 |
ENSMUST00000216811.2
|
Olfr68
|
olfactory receptor 68 |
| chrX_+_9751861 | 0.15 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr6_-_144978557 | 0.15 |
ENSMUST00000136819.3
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr8_+_85065268 | 0.15 |
ENSMUST00000238701.2
|
Cacna1a
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr7_+_44805280 | 0.15 |
ENSMUST00000107811.4
ENSMUST00000211414.2 |
Pih1d1
|
PIH1 domain containing 1 |
| chr19_-_46661321 | 0.15 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_-_99134885 | 0.15 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr1_+_179928709 | 0.15 |
ENSMUST00000133890.8
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr10_+_29019645 | 0.15 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chrX_-_72080709 | 0.14 |
ENSMUST00000114540.4
ENSMUST00000051569.7 |
Pnma5
|
paraneoplastic antigen family 5 |
| chr8_-_94825556 | 0.14 |
ENSMUST00000034206.6
|
Bbs2
|
Bardet-Biedl syndrome 2 (human) |
| chr19_+_53781721 | 0.14 |
ENSMUST00000162910.2
|
Rbm20
|
RNA binding motif protein 20 |
| chr7_-_16019935 | 0.14 |
ENSMUST00000145519.3
|
Ccdc9
|
coiled-coil domain containing 9 |
| chr2_-_86070633 | 0.14 |
ENSMUST00000215607.3
|
Olfr1048
|
olfactory receptor 1048 |
| chr9_+_96140781 | 0.14 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr13_+_43276323 | 0.14 |
ENSMUST00000136576.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr18_-_32170012 | 0.14 |
ENSMUST00000134663.2
|
Myo7b
|
myosin VIIB |
| chr9_-_58109627 | 0.14 |
ENSMUST00000216231.2
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr11_+_92989229 | 0.14 |
ENSMUST00000107859.8
ENSMUST00000107861.8 ENSMUST00000042943.13 ENSMUST00000107858.9 |
Car10
|
carbonic anhydrase 10 |
| chr4_+_52596266 | 0.14 |
ENSMUST00000029995.6
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr6_+_96092230 | 0.14 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr19_+_13208692 | 0.14 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr11_+_66802807 | 0.14 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr7_-_19297001 | 0.14 |
ENSMUST00000058444.10
|
Ppp1r37
|
protein phosphatase 1, regulatory subunit 37 |
| chr7_+_103197281 | 0.14 |
ENSMUST00000214173.2
|
Olfr613
|
olfactory receptor 613 |
| chr3_-_85909798 | 0.14 |
ENSMUST00000061343.4
|
Prss48
|
protease, serine 48 |
| chr3_-_92346078 | 0.14 |
ENSMUST00000062160.4
|
Sprr1b
|
small proline-rich protein 1B |
| chr5_-_107073704 | 0.14 |
ENSMUST00000200249.2
ENSMUST00000112690.10 |
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr5_-_137305895 | 0.14 |
ENSMUST00000199243.5
ENSMUST00000197466.5 ENSMUST00000040873.12 |
Srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr2_-_120075639 | 0.14 |
ENSMUST00000090071.5
|
Pla2g4e
|
phospholipase A2, group IVE |
| chr10_-_129107354 | 0.14 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr7_-_103191732 | 0.14 |
ENSMUST00000215663.2
|
Olfr612
|
olfactory receptor 612 |
| chr1_+_40468720 | 0.13 |
ENSMUST00000174335.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr13_-_32960379 | 0.13 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr9_+_19404591 | 0.13 |
ENSMUST00000214130.2
|
Olfr851
|
olfactory receptor 851 |
| chr9_-_58108988 | 0.13 |
ENSMUST00000163200.3
ENSMUST00000165276.2 ENSMUST00000214647.2 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr7_-_98831916 | 0.13 |
ENSMUST00000033001.6
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr9_-_58109526 | 0.13 |
ENSMUST00000216297.2
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr4_-_119047180 | 0.13 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr5_-_87638728 | 0.13 |
ENSMUST00000147854.6
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr19_+_12647803 | 0.13 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr5_+_104447037 | 0.13 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr10_+_129084281 | 0.13 |
ENSMUST00000214109.2
|
Olfr775
|
olfactory receptor 775 |
| chr15_+_99590098 | 0.13 |
ENSMUST00000228185.2
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr2_+_176696346 | 0.13 |
ENSMUST00000126358.8
|
Gm14419
|
predicted gene 14419 |
| chr2_+_85715984 | 0.13 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr12_-_73093953 | 0.13 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr2_+_176403726 | 0.13 |
ENSMUST00000108983.4
|
Gm14305
|
predicted gene 14305 |
| chr11_+_92990110 | 0.13 |
ENSMUST00000107863.4
|
Car10
|
carbonic anhydrase 10 |
| chr1_-_165830160 | 0.13 |
ENSMUST00000111429.11
ENSMUST00000176800.2 ENSMUST00000177358.8 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr7_-_85895409 | 0.13 |
ENSMUST00000165771.2
ENSMUST00000233075.2 ENSMUST00000233317.2 ENSMUST00000233312.2 ENSMUST00000233928.2 ENSMUST00000232799.2 ENSMUST00000233744.2 |
Vmn2r76
|
vomeronasal 2, receptor 76 |
| chr6_+_65567373 | 0.13 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr14_+_52962756 | 0.13 |
ENSMUST00000181483.3
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr9_+_54446268 | 0.13 |
ENSMUST00000060242.12
|
Sh2d7
|
SH2 domain containing 7 |
| chr7_-_86425016 | 0.13 |
ENSMUST00000107271.10
|
Folh1
|
folate hydrolase 1 |
| chr4_+_152423075 | 0.13 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chrX_-_105528503 | 0.13 |
ENSMUST00000138724.8
ENSMUST00000149331.2 |
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr6_+_56979285 | 0.12 |
ENSMUST00000079669.7
|
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr6_-_66537080 | 0.12 |
ENSMUST00000079584.3
ENSMUST00000227014.2 |
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr1_-_38875757 | 0.12 |
ENSMUST00000147695.9
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr3_-_58729732 | 0.12 |
ENSMUST00000191233.4
|
Mindy4b-ps
|
MINDY lysine 48 deubiquitinase 4B, pseudogene |
| chr1_+_106908709 | 0.12 |
ENSMUST00000027564.8
|
Serpinb13
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr5_+_27109679 | 0.12 |
ENSMUST00000120555.8
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr2_-_77349909 | 0.12 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr6_-_130003917 | 0.12 |
ENSMUST00000074056.3
|
Klra6
|
killer cell lectin-like receptor, subfamily A, member 6 |
| chr5_-_86666408 | 0.12 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr16_+_14523696 | 0.12 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr7_-_103094646 | 0.12 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr9_+_38202403 | 0.12 |
ENSMUST00000216276.2
ENSMUST00000215219.2 |
Olfr896-ps1
|
olfactory receptor 896, pseudogene 1 |
| chr6_+_21215472 | 0.12 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr18_+_20643325 | 0.12 |
ENSMUST00000070892.8
ENSMUST00000234945.2 |
Dsg3
|
desmoglein 3 |
| chr16_-_26345493 | 0.12 |
ENSMUST00000165687.3
|
Tmem207
|
transmembrane protein 207 |
| chr6_+_90099578 | 0.12 |
ENSMUST00000227766.2
ENSMUST00000227109.2 ENSMUST00000226977.2 ENSMUST00000228880.2 ENSMUST00000174204.3 ENSMUST00000227911.2 ENSMUST00000226542.2 ENSMUST00000227876.2 |
Vmn1r51
|
vomeronasal 1 receptor 51 |
| chr11_-_58353096 | 0.12 |
ENSMUST00000215691.2
|
Olfr30
|
olfactory receptor 30 |
| chr7_-_86425147 | 0.12 |
ENSMUST00000001824.7
|
Folh1
|
folate hydrolase 1 |
| chrX_+_111524998 | 0.12 |
ENSMUST00000122805.2
|
Zfp711
|
zinc finger protein 711 |
| chr17_+_28532487 | 0.12 |
ENSMUST00000114803.9
ENSMUST00000114801.9 ENSMUST00000114804.11 |
Fance
|
Fanconi anemia, complementation group E |
| chr6_+_134988572 | 0.12 |
ENSMUST00000032326.11
ENSMUST00000130851.8 ENSMUST00000205244.3 ENSMUST00000205055.3 ENSMUST00000204646.3 ENSMUST00000154558.3 |
Ddx47
|
DEAD box helicase 47 |
| chr5_-_104225458 | 0.12 |
ENSMUST00000198485.5
ENSMUST00000164471.8 ENSMUST00000178967.2 |
Gm17660
|
predicted gene, 17660 |
| chr9_-_19799300 | 0.12 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chr5_-_120750623 | 0.12 |
ENSMUST00000140554.2
ENSMUST00000031599.9 ENSMUST00000177800.8 |
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr19_-_33728759 | 0.11 |
ENSMUST00000147153.4
|
Lipo2
|
lipase, member O2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.3 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 1.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:0051542 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.2 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.1 | 0.2 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.1 | 0.1 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.3 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.6 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.4 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.6 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0021750 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0048880 | proprioception(GO:0019230) sensory system development(GO:0048880) |
| 0.0 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.8 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.2 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.1 | GO:0015898 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:2000397 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.2 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 1.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 1.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0090158 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.0 | 0.1 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 2.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.2 | 0.6 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 3.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0052597 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.0 | 0.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 1.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 3.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |