Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
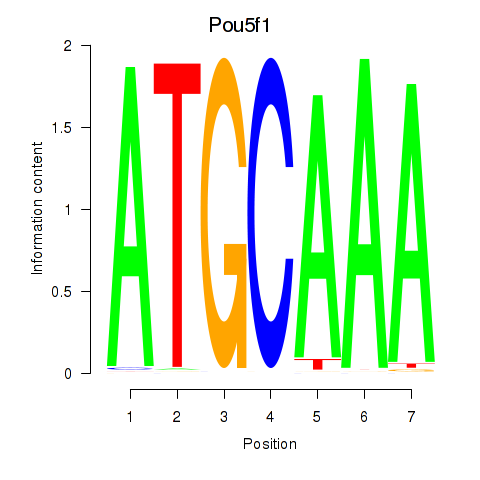
Results for Pou5f1
Z-value: 2.13
Transcription factors associated with Pou5f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou5f1
|
ENSMUSG00000024406.17 | Pou5f1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou5f1 | mm39_v1_chr17_+_35816915_35816968 | 0.09 | 5.8e-01 | Click! |
Activity profile of Pou5f1 motif
Sorted Z-values of Pou5f1 motif
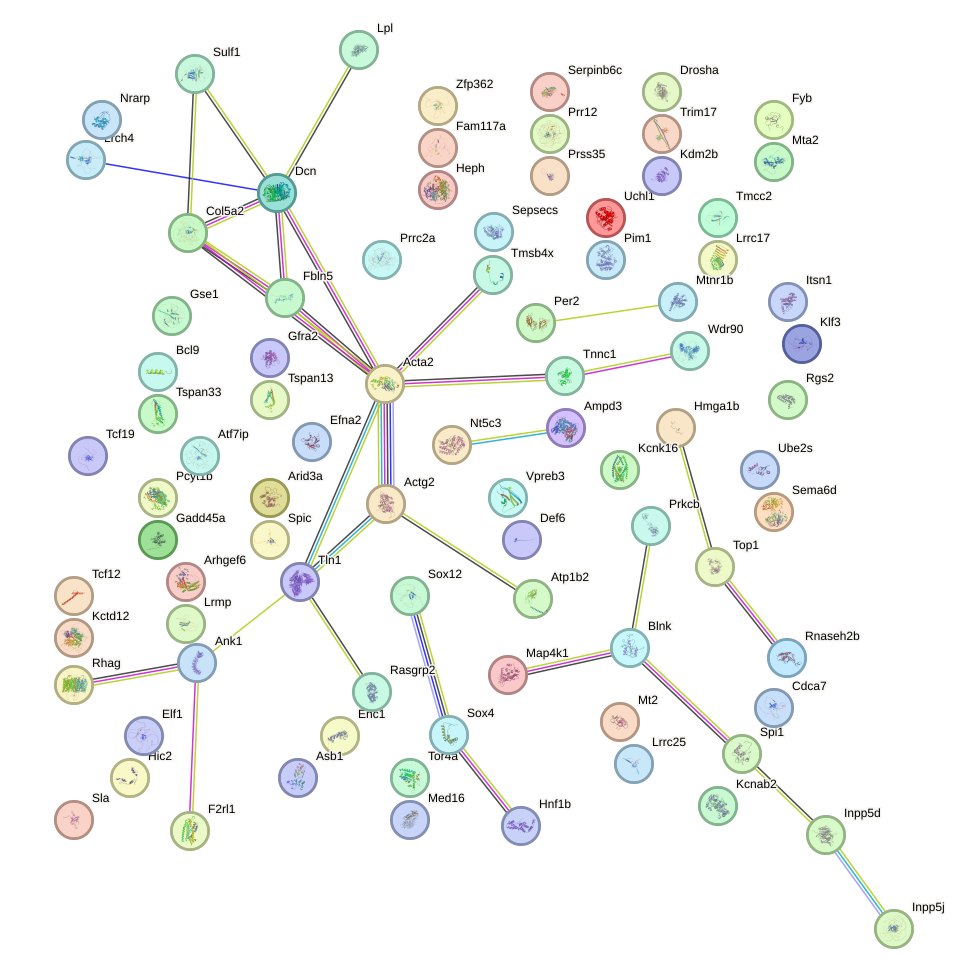
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou5f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_69496655 | 14.01 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr10_+_75784126 | 9.00 |
ENSMUST00000000926.3
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chr17_+_41121979 | 8.61 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr6_-_67014383 | 8.21 |
ENSMUST00000043098.9
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr11_-_106205320 | 7.58 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr8_+_94899292 | 7.40 |
ENSMUST00000034214.8
ENSMUST00000212806.2 |
Mt2
|
metallothionein 2 |
| chr11_+_95227836 | 6.81 |
ENSMUST00000037502.7
|
Fam117a
|
family with sequence similarity 117, member A |
| chr1_-_132318039 | 6.70 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr7_-_4815542 | 6.17 |
ENSMUST00000079496.9
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr17_+_28426752 | 5.95 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr7_+_28682253 | 5.64 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr17_+_27775637 | 5.56 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr6_-_83513222 | 5.48 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr17_+_27775613 | 5.43 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_27775471 | 5.41 |
ENSMUST00000118599.9
ENSMUST00000232265.2 ENSMUST00000232013.2 ENSMUST00000114888.11 ENSMUST00000231874.2 ENSMUST00000119486.9 ENSMUST00000231825.2 ENSMUST00000231866.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr12_-_36092475 | 5.21 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr4_-_152561896 | 4.76 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr15_-_66703471 | 4.74 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr6_-_67014348 | 4.67 |
ENSMUST00000204369.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr6_-_68713748 | 4.52 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr6_+_29694181 | 4.52 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr11_-_115968745 | 4.43 |
ENSMUST00000156545.2
ENSMUST00000075036.9 ENSMUST00000106451.8 |
Unc13d
|
unc-13 homolog D |
| chr1_-_45542442 | 4.38 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr1_+_91468409 | 4.26 |
ENSMUST00000027538.9
ENSMUST00000190484.7 ENSMUST00000186068.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr12_-_113542610 | 4.23 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr2_-_152239966 | 4.23 |
ENSMUST00000063332.9
ENSMUST00000182625.2 |
Sox12
|
SRY (sex determining region Y)-box 12 |
| chr8_+_69333143 | 4.19 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr4_-_63965161 | 4.15 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr6_+_145067457 | 4.09 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr4_+_152123772 | 4.00 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr8_+_23464860 | 3.91 |
ENSMUST00000110688.9
ENSMUST00000121802.9 |
Ank1
|
ankyrin 1, erythroid |
| chr10_+_115653152 | 3.84 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr4_-_128699838 | 3.76 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr14_-_20319242 | 3.69 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr6_-_67014191 | 3.64 |
ENSMUST00000204282.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr6_-_56878854 | 3.61 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr10_+_79762858 | 3.58 |
ENSMUST00000019708.12
ENSMUST00000105377.8 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr2_+_90927053 | 3.50 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr8_+_23548541 | 3.45 |
ENSMUST00000173248.8
|
Ank1
|
ankyrin 1, erythroid |
| chr8_+_71069476 | 3.41 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr6_-_83513184 | 3.37 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr12_-_113552322 | 3.36 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chr17_-_35827676 | 3.28 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr1_+_87603952 | 3.24 |
ENSMUST00000170300.8
ENSMUST00000167032.2 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr19_-_34231600 | 3.24 |
ENSMUST00000238147.2
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr7_-_44180700 | 3.17 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr11_+_44508137 | 3.15 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr10_-_88520877 | 3.15 |
ENSMUST00000138734.2
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr13_-_22017677 | 3.07 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr14_-_67953035 | 3.06 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr6_+_68247469 | 3.01 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr19_+_8919228 | 3.00 |
ENSMUST00000096240.3
|
Mta2
|
metastasis-associated gene family, member 2 |
| chr12_-_113589576 | 3.00 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr7_+_110372860 | 2.99 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr13_-_29137673 | 2.94 |
ENSMUST00000067230.6
|
Sox4
|
SRY (sex determining region Y)-box 4 |
| chr19_+_6450553 | 2.92 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr11_-_115968576 | 2.91 |
ENSMUST00000106450.8
|
Unc13d
|
unc-13 homolog D |
| chr11_+_98632631 | 2.89 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr14_+_71127540 | 2.87 |
ENSMUST00000022699.10
|
Gfra2
|
glial cell line derived neurotrophic factor family receptor alpha 2 |
| chr5_+_137628377 | 2.86 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr5_+_137627431 | 2.85 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr6_+_67586695 | 2.85 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr12_-_113823290 | 2.85 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr2_+_72306503 | 2.83 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chrX_+_92718695 | 2.83 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr12_-_114252202 | 2.75 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr17_+_28426831 | 2.72 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6 |
| chr7_+_121888520 | 2.71 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr5_+_76988444 | 2.70 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr11_+_58845502 | 2.70 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr11_-_3454766 | 2.66 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr12_-_114443071 | 2.65 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr17_-_35383867 | 2.65 |
ENSMUST00000025253.12
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr13_+_97377604 | 2.63 |
ENSMUST00000041623.9
|
Enc1
|
ectodermal-neural cortex 1 |
| chr12_-_113561594 | 2.62 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr5_+_66833434 | 2.62 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr1_+_91468266 | 2.59 |
ENSMUST00000086843.11
|
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr11_+_83741689 | 2.58 |
ENSMUST00000108114.9
|
Hnf1b
|
HNF1 homeobox B |
| chr11_+_83741657 | 2.54 |
ENSMUST00000021016.10
|
Hnf1b
|
HNF1 homeobox B |
| chr12_-_101784727 | 2.51 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr17_+_29709723 | 2.41 |
ENSMUST00000024811.9
|
Pim1
|
proviral integration site 1 |
| chr16_+_17051423 | 2.34 |
ENSMUST00000090190.14
ENSMUST00000232082.2 ENSMUST00000232426.2 |
Hic2
Gm49573
|
hypermethylated in cancer 2 predicted gene, 49573 |
| chr6_-_69792108 | 2.33 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr5_-_123126550 | 2.28 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr14_-_103220066 | 2.16 |
ENSMUST00000184744.2
|
Kctd12
|
potassium channel tetramerisation domain containing 12 |
| chr7_+_110373447 | 2.15 |
ENSMUST00000147587.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr6_+_68233361 | 2.14 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr11_+_98632696 | 2.14 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr10_-_79744726 | 2.13 |
ENSMUST00000165684.8
ENSMUST00000164705.8 ENSMUST00000105378.9 ENSMUST00000170409.2 |
Med16
|
mediator complex subunit 16 |
| chr6_-_70094604 | 2.11 |
ENSMUST00000103378.3
|
Igkv8-30
|
immunoglobulin kappa chain variable 8-30 |
| chr6_+_136495784 | 2.10 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr12_-_101785307 | 2.09 |
ENSMUST00000021603.9
|
Fbln5
|
fibulin 5 |
| chr11_-_115968373 | 2.08 |
ENSMUST00000174822.8
|
Unc13d
|
unc-13 homolog D |
| chr2_-_25086810 | 2.08 |
ENSMUST00000081869.7
|
Tor4a
|
torsin family 4, member A |
| chr5_-_123127148 | 2.06 |
ENSMUST00000046073.16
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr2_+_160487801 | 2.05 |
ENSMUST00000109468.3
|
Top1
|
topoisomerase (DNA) I |
| chr10_-_117212860 | 2.04 |
ENSMUST00000069168.13
ENSMUST00000176686.8 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr17_-_28705055 | 2.04 |
ENSMUST00000233870.2
|
Fkbp5
|
FK506 binding protein 5 |
| chr4_-_43562397 | 2.03 |
ENSMUST00000030187.14
|
Tln1
|
talin 1 |
| chr7_-_44702269 | 2.01 |
ENSMUST00000057293.8
|
Prr12
|
proline rich 12 |
| chrX_-_165992311 | 2.01 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr1_-_143879877 | 2.00 |
ENSMUST00000127206.8
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr6_+_68098030 | 1.99 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr12_-_114355789 | 1.99 |
ENSMUST00000103486.2
|
Ighv6-3
|
immunoglobulin heavy variable 6-3 |
| chr17_-_28705082 | 1.98 |
ENSMUST00000079413.11
|
Fkbp5
|
FK506 binding protein 5 |
| chr12_-_114321838 | 1.98 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr3_+_5815863 | 1.97 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr2_+_25070749 | 1.95 |
ENSMUST00000104999.4
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr5_+_64969679 | 1.94 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr12_-_113649535 | 1.92 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr14_+_71127882 | 1.91 |
ENSMUST00000227697.2
|
Gfra2
|
glial cell line derived neurotrophic factor family receptor alpha 2 |
| chr9_+_86625694 | 1.90 |
ENSMUST00000179574.2
ENSMUST00000036426.13 |
Prss35
|
protease, serine 35 |
| chr11_+_98632953 | 1.90 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr14_+_62529924 | 1.89 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chrX_-_165992145 | 1.88 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr16_+_91526169 | 1.87 |
ENSMUST00000114001.8
ENSMUST00000113999.8 ENSMUST00000064797.12 ENSMUST00000114002.9 ENSMUST00000095909.10 ENSMUST00000056482.14 ENSMUST00000113996.8 |
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr15_+_12824925 | 1.86 |
ENSMUST00000090292.13
|
Drosha
|
drosha, ribonuclease type III |
| chr13_-_22227114 | 1.85 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr12_-_113860566 | 1.85 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr9_-_71803354 | 1.84 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr12_-_114398864 | 1.83 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr15_+_6609322 | 1.81 |
ENSMUST00000090461.12
|
Fyb
|
FYN binding protein |
| chr12_-_114752425 | 1.79 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr10_+_97315465 | 1.77 |
ENSMUST00000105287.11
|
Dcn
|
decorin |
| chr6_-_69741999 | 1.76 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr8_+_121215155 | 1.75 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr2_+_124452493 | 1.74 |
ENSMUST00000103239.10
ENSMUST00000103240.9 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr12_-_114487525 | 1.72 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr1_+_12788720 | 1.70 |
ENSMUST00000088585.10
|
Sulf1
|
sulfatase 1 |
| chr6_+_68026941 | 1.68 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr15_+_12824901 | 1.65 |
ENSMUST00000169061.8
|
Drosha
|
drosha, ribonuclease type III |
| chr12_-_113666198 | 1.62 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr1_-_91386976 | 1.58 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2 |
| chr17_+_28059036 | 1.56 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr4_+_128582519 | 1.55 |
ENSMUST00000106080.8
|
Phc2
|
polyhomeotic 2 |
| chr13_-_95661726 | 1.55 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr10_+_79763164 | 1.52 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chrX_-_56384089 | 1.50 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr2_-_25086770 | 1.50 |
ENSMUST00000142857.2
ENSMUST00000137920.2 |
Tor4a
|
torsin family 4, member A |
| chr19_-_40982576 | 1.49 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr12_-_113575237 | 1.49 |
ENSMUST00000178229.3
|
Ighv2-3
|
immunoglobulin heavy variable 2-3 |
| chr6_-_70237939 | 1.48 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr6_-_70292451 | 1.47 |
ENSMUST00000103387.3
|
Igkv8-21
|
immunoglobulin kappa variable 8-21 |
| chr12_-_113790741 | 1.46 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr11_-_5787743 | 1.46 |
ENSMUST00000109837.8
|
Polm
|
polymerase (DNA directed), mu |
| chr6_+_68279392 | 1.45 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr9_-_85209162 | 1.45 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr1_-_143879738 | 1.43 |
ENSMUST00000153527.3
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr19_+_6450641 | 1.42 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr5_+_21748523 | 1.41 |
ENSMUST00000035651.6
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr13_+_22227359 | 1.37 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr6_-_69282389 | 1.37 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr9_-_85209340 | 1.36 |
ENSMUST00000187711.2
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr3_+_96177010 | 1.36 |
ENSMUST00000051089.4
ENSMUST00000177113.2 |
Gm42743
H2bc18
|
predicted gene 42743 H2B clustered histone 18 |
| chr3_-_97134680 | 1.33 |
ENSMUST00000046521.14
|
Bcl9
|
B cell CLL/lymphoma 9 |
| chr12_-_113802603 | 1.32 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr14_+_79753055 | 1.31 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr12_-_114117264 | 1.31 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr12_-_115258124 | 1.30 |
ENSMUST00000192591.2
|
Ighv8-8
|
immunoglobulin heavy variable 8-8 |
| chr12_-_114621406 | 1.30 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr12_-_113733922 | 1.28 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr8_+_55407872 | 1.26 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr18_+_36780913 | 1.25 |
ENSMUST00000140061.8
|
Maskbp3
|
multiple ankyrin repeats single KH domain binding protein 3 |
| chr6_+_70348416 | 1.25 |
ENSMUST00000103391.4
|
Igkv6-17
|
immunoglobulin kappa variable 6-17 |
| chr8_-_116434517 | 1.22 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr6_-_70383976 | 1.19 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr12_-_113912416 | 1.13 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr12_-_114672701 | 1.13 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr6_-_70318437 | 1.13 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr17_+_27276262 | 1.11 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr4_+_3678108 | 1.09 |
ENSMUST00000041377.13
ENSMUST00000103010.4 |
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr12_-_114226570 | 1.09 |
ENSMUST00000103479.4
ENSMUST00000195619.2 |
Ighv3-5
|
immunoglobulin heavy variable 3-5 |
| chr6_-_29216276 | 1.08 |
ENSMUST00000162215.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr6_+_68657317 | 1.07 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr16_-_4698148 | 1.06 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr10_+_119655294 | 1.06 |
ENSMUST00000105262.9
ENSMUST00000147454.8 ENSMUST00000138410.8 ENSMUST00000144825.8 ENSMUST00000148954.8 ENSMUST00000144959.8 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr16_+_23043474 | 1.06 |
ENSMUST00000023601.14
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr8_+_31640332 | 1.05 |
ENSMUST00000209851.2
ENSMUST00000098842.3 ENSMUST00000210129.2 |
Tti2
|
TELO2 interacting protein 2 |
| chr6_-_70149254 | 1.05 |
ENSMUST00000197272.2
|
Igkv8-27
|
immunoglobulin kappa chain variable 8-27 |
| chr7_+_24981604 | 1.04 |
ENSMUST00000163320.8
ENSMUST00000005578.13 |
Cic
|
capicua transcriptional repressor |
| chr2_+_28083105 | 1.03 |
ENSMUST00000100244.10
|
Olfm1
|
olfactomedin 1 |
| chr7_-_15781838 | 1.03 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr9_-_57168777 | 1.02 |
ENSMUST00000217657.2
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr11_+_115272732 | 1.01 |
ENSMUST00000053288.6
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr12_-_115172211 | 1.00 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr6_-_70364222 | 0.99 |
ENSMUST00000103392.3
ENSMUST00000195945.2 |
Igkv8-16
|
immunoglobulin kappa variable 8-16 |
| chr5_-_115791032 | 0.99 |
ENSMUST00000121746.8
ENSMUST00000118576.8 |
Bicdl1
|
BICD family like cargo adaptor 1 |
| chr13_+_21906214 | 0.98 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr7_-_28071919 | 0.98 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr6_-_70194405 | 0.98 |
ENSMUST00000103384.2
|
Igkv8-24
|
immunoglobulin kappa chain variable 8-24 |
| chr7_-_37472979 | 0.97 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr9_+_75093177 | 0.97 |
ENSMUST00000129281.8
ENSMUST00000148144.8 ENSMUST00000130384.2 |
Myo5a
|
myosin VA |
| chr8_-_116433733 | 0.96 |
ENSMUST00000069009.7
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr10_+_128542120 | 0.96 |
ENSMUST00000054125.9
|
Pmel
|
premelanosome protein |
| chr7_-_125799644 | 0.95 |
ENSMUST00000168189.8
|
Xpo6
|
exportin 6 |
| chr5_+_75316552 | 0.94 |
ENSMUST00000168162.5
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr3_+_75982890 | 0.94 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr8_-_65489834 | 0.93 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr13_+_23930717 | 0.93 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr13_+_5911481 | 0.92 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 14.0 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 2.2 | 8.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.7 | 6.9 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.3 | 4.0 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 1.3 | 5.1 | GO:0061215 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.2 | 6.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.2 | 16.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.2 | 9.4 | GO:0002432 | granuloma formation(GO:0002432) |
| 1.1 | 4.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 1.0 | 4.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.0 | 4.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.9 | 7.4 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.9 | 9.0 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.8 | 3.2 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.7 | 3.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.7 | 4.6 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.6 | 4.2 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.6 | 2.9 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.5 | 2.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.5 | 1.6 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.5 | 1.6 | GO:0070948 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.5 | 5.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 3.4 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.4 | 1.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 1.3 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.4 | 3.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.4 | 2.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 0.8 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.4 | 1.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.4 | 3.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 48.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 5.7 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 0.9 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 2.7 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.3 | 16.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.3 | 1.7 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 0.5 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) |
| 0.3 | 0.8 | GO:2000040 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.3 | 4.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 2.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 3.9 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 2.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 0.9 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.2 | 2.4 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.2 | 1.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 4.8 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.2 | 3.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.0 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 0.8 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 1.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.2 | 1.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 3.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 0.7 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.2 | 7.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 0.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.8 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 1.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 2.0 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 1.5 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 4.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.2 | GO:2000471 | regulation of eosinophil degranulation(GO:0043309) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 2.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 8.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 1.9 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.1 | 2.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.5 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.7 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 | 22.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 6.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 1.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.6 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 0.2 | GO:0071336 | hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.4 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 4.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.5 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 1.0 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.4 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 4.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.7 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 1.3 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 1.3 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 3.1 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 3.1 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 1.5 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 3.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 2.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 2.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.2 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.2 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.0 | 1.6 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 2.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0060762 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 1.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 4.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 2.3 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 1.7 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.9 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 2.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 16.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.6 | 4.8 | GO:1990031 | pinceau fiber(GO:1990031) |
| 1.5 | 4.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.4 | 14.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.2 | 3.5 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.9 | 9.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.9 | 4.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.8 | 7.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 48.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 4.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.4 | 3.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 1.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 2.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 1.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.3 | 7.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 1.0 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 2.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 6.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 4.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.8 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 4.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.6 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 1.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 2.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.8 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 4.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 19.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 5.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 5.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 2.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 6.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 2.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 4.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 4.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 3.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 3.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 5.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 21.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 16.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.7 | 6.9 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.2 | 14.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.0 | 4.8 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.9 | 2.7 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.7 | 7.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.7 | 2.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.7 | 3.5 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.7 | 2.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.7 | 2.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.6 | 4.0 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.6 | 12.3 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.5 | 5.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 3.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.4 | 4.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 1.6 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.4 | 1.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 48.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 5.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 0.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 0.8 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.3 | 1.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 1.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 1.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 2.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 2.7 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.2 | 1.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 3.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 0.7 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.2 | 4.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 0.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 4.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 0.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 2.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 1.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 1.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 0.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 3.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 3.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 2.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 6.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 4.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 6.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 18.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 5.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.1 | 3.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 3.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 4.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 1.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 6.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 4.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 3.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 2.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 15.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 3.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 21.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 10.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 8.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 7.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 4.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 9.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 6.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 3.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 16.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 13.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 13.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 12.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 8.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 5.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 3.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 7.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 9.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 4.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 6.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 13.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 4.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |