Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
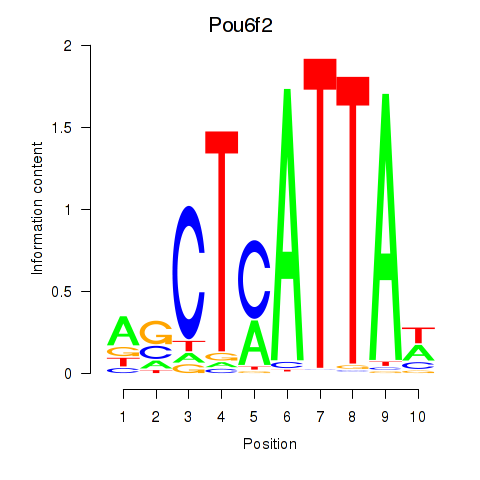
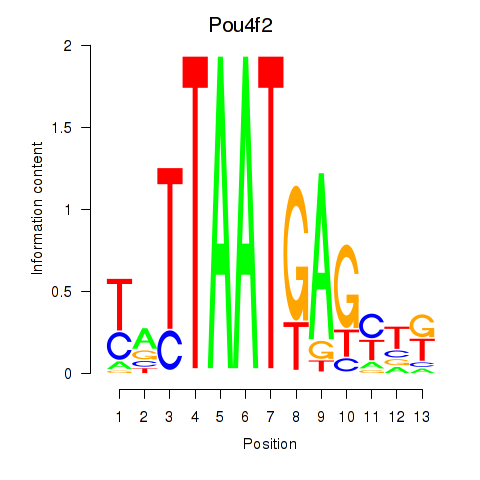
Results for Pou6f2_Pou4f2
Z-value: 0.80
Transcription factors associated with Pou6f2_Pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou6f2
|
ENSMUSG00000009734.19 | Pou6f2 |
|
Pou4f2
|
ENSMUSG00000031688.5 | Pou4f2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou6f2 | mm39_v1_chr13_-_18556626_18556626 | 0.04 | 8.2e-01 | Click! |
Activity profile of Pou6f2_Pou4f2 motif
Sorted Z-values of Pou6f2_Pou4f2 motif
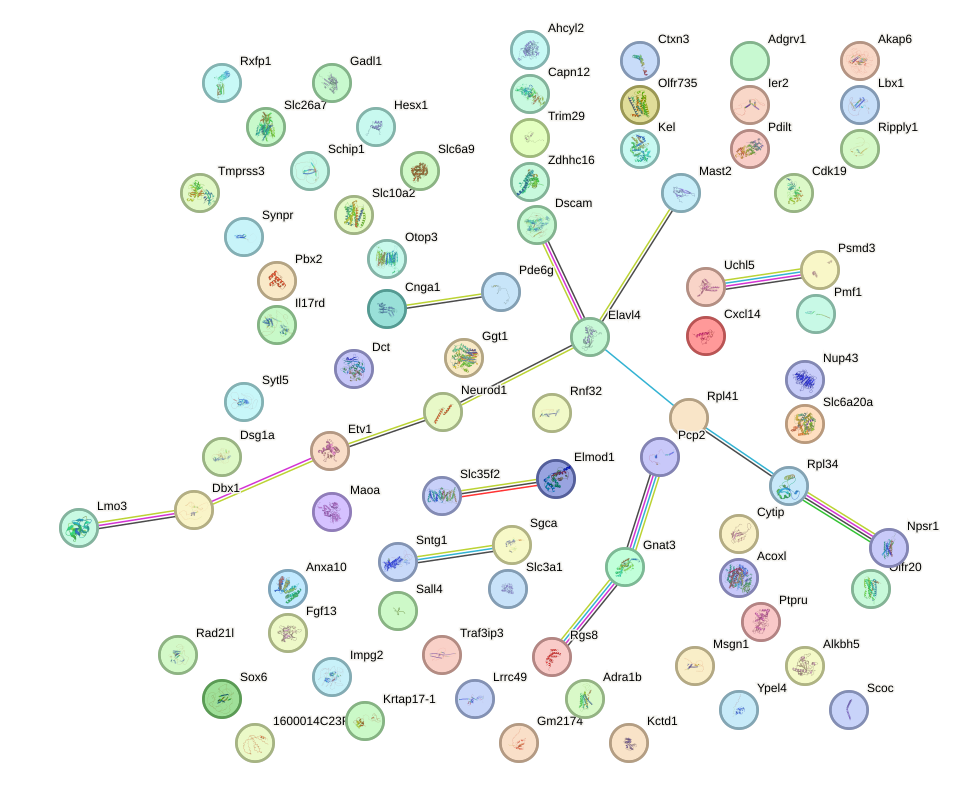
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou6f2_Pou4f2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_75402090 | 1.59 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chrX_+_55493325 | 1.39 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr6_-_41681273 | 1.35 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr13_-_56444118 | 1.26 |
ENSMUST00000224801.2
|
Cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr4_+_117706559 | 1.17 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr17_+_85335775 | 1.09 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr4_+_117706390 | 1.08 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr8_-_5155347 | 0.86 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr2_+_84564394 | 0.62 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr1_+_143653001 | 0.61 |
ENSMUST00000189936.7
ENSMUST00000018333.13 ENSMUST00000185493.7 |
Uchl5
|
ubiquitin carboxyl-terminal esterase L5 |
| chr14_+_26722319 | 0.51 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr14_-_118289557 | 0.51 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr12_-_25147139 | 0.50 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr7_-_115630282 | 0.49 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_153079828 | 0.47 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr9_+_115738523 | 0.42 |
ENSMUST00000119291.8
ENSMUST00000069651.13 |
Gadl1
|
glutamate decarboxylase-like 1 |
| chr3_-_130524024 | 0.41 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr4_-_58499398 | 0.38 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr10_-_128384971 | 0.38 |
ENSMUST00000176906.2
|
Rpl41
|
ribosomal protein L41 |
| chr10_-_128384994 | 0.37 |
ENSMUST00000177163.8
ENSMUST00000176683.8 ENSMUST00000176010.8 |
Rpl41
|
ribosomal protein L41 |
| chr2_-_168608949 | 0.37 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr12_-_11258973 | 0.36 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr3_-_130523954 | 0.33 |
ENSMUST00000196202.5
ENSMUST00000133802.6 ENSMUST00000062601.14 ENSMUST00000200517.2 |
Rpl34
|
ribosomal protein L34 |
| chr4_-_14621805 | 0.32 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr13_-_81718759 | 0.32 |
ENSMUST00000109565.9
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr12_+_52746158 | 0.32 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr2_+_37101586 | 0.32 |
ENSMUST00000214897.2
|
Olfr366
|
olfactory receptor 366 |
| chr2_-_58050494 | 0.30 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr8_-_3675274 | 0.30 |
ENSMUST00000004749.7
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr2_-_17465410 | 0.29 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr4_-_14621669 | 0.28 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr9_-_123507847 | 0.28 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr7_-_119122681 | 0.27 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr10_+_40225272 | 0.27 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr8_-_68270870 | 0.26 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_153541020 | 0.26 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr3_-_88317601 | 0.26 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr11_+_115225557 | 0.25 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr14_-_9015639 | 0.23 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chrX_-_138683102 | 0.23 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr2_-_168609110 | 0.22 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr8_-_62576140 | 0.22 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr1_-_192880260 | 0.21 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr4_-_116228921 | 0.19 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr9_+_53678801 | 0.18 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr6_-_138399896 | 0.18 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr4_-_110144676 | 0.18 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr2_-_79287095 | 0.17 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr16_+_43323970 | 0.17 |
ENSMUST00000126100.8
ENSMUST00000123047.8 ENSMUST00000156981.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr18_-_15197138 | 0.17 |
ENSMUST00000234864.2
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr15_-_50746202 | 0.17 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr6_+_29859372 | 0.17 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr3_+_68479578 | 0.17 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr10_+_7543260 | 0.16 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr7_+_28580847 | 0.16 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr11_-_99884818 | 0.15 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr4_-_116263183 | 0.15 |
ENSMUST00000123072.8
ENSMUST00000144281.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr14_+_26761023 | 0.15 |
ENSMUST00000223942.2
|
Il17rd
|
interleukin 17 receptor D |
| chr19_+_41921903 | 0.15 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chrX_+_16485937 | 0.15 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr5_+_18167547 | 0.15 |
ENSMUST00000030561.9
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr6_+_106095726 | 0.15 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr11_-_94864273 | 0.15 |
ENSMUST00000100551.11
ENSMUST00000152042.2 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr2_-_111843053 | 0.15 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr9_-_53882530 | 0.15 |
ENSMUST00000048409.14
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr5_+_29400981 | 0.14 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr8_-_84184978 | 0.14 |
ENSMUST00000081506.11
|
Scoc
|
short coiled-coil protein |
| chrX_+_9751861 | 0.14 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr9_+_24194729 | 0.13 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr11_-_120344299 | 0.13 |
ENSMUST00000026452.3
|
Pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr7_-_49286594 | 0.13 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr2_+_127750978 | 0.13 |
ENSMUST00000110344.2
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr6_+_29859660 | 0.13 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr4_-_14621497 | 0.13 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr11_-_43727071 | 0.13 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr2_-_88157559 | 0.13 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr8_-_3675024 | 0.13 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr8_-_3674993 | 0.12 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr6_+_29859685 | 0.12 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr2_+_88644840 | 0.12 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr16_-_96971905 | 0.12 |
ENSMUST00000056102.9
|
Dscam
|
DS cell adhesion molecule |
| chr16_+_56024676 | 0.12 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr10_+_73782857 | 0.12 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr17_+_34811217 | 0.11 |
ENSMUST00000038149.13
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr6_-_41752111 | 0.11 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chrX_-_58211440 | 0.11 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr7_-_102566717 | 0.11 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr19_+_55730696 | 0.11 |
ENSMUST00000153888.9
ENSMUST00000127233.9 ENSMUST00000061496.17 ENSMUST00000111656.8 ENSMUST00000111657.11 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr11_+_73851643 | 0.10 |
ENSMUST00000213134.2
ENSMUST00000216291.2 |
Olfr397
|
olfactory receptor 397 |
| chr12_+_38831093 | 0.10 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr18_+_20443795 | 0.10 |
ENSMUST00000077146.4
|
Dsg1a
|
desmoglein 1 alpha |
| chr10_+_102348076 | 0.09 |
ENSMUST00000219445.2
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr5_-_72800070 | 0.09 |
ENSMUST00000087213.12
|
Cnga1
|
cyclic nucleotide gated channel alpha 1 |
| chr16_+_43067641 | 0.09 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_9368721 | 0.09 |
ENSMUST00000132064.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr9_-_60557076 | 0.09 |
ENSMUST00000053171.14
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr5_+_139529643 | 0.09 |
ENSMUST00000174792.2
|
Uncx
|
UNC homeobox |
| chr3_-_79645101 | 0.09 |
ENSMUST00000078527.13
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr17_-_31417932 | 0.09 |
ENSMUST00000114549.4
|
Tmprss3
|
transmembrane protease, serine 3 |
| chr11_+_60428788 | 0.09 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr17_-_46044768 | 0.08 |
ENSMUST00000178179.2
|
1600014C23Rik
|
RIKEN cDNA 1600014C23 gene |
| chr18_+_57601541 | 0.08 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr9_+_43222104 | 0.08 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr12_+_38830812 | 0.08 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr2_-_151510453 | 0.08 |
ENSMUST00000180195.8
ENSMUST00000096439.4 |
Rad21l
|
RAD21-like (S. pombe) |
| chr8_-_85389470 | 0.08 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr14_-_50586329 | 0.08 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr19_-_45224251 | 0.08 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr17_-_37399343 | 0.08 |
ENSMUST00000207101.2
ENSMUST00000217397.2 ENSMUST00000215195.2 ENSMUST00000216488.2 |
Olfr90
|
olfactory receptor 90 |
| chr2_+_85835884 | 0.08 |
ENSMUST00000111589.3
|
Olfr1032
|
olfactory receptor 1032 |
| chrX_+_110808231 | 0.08 |
ENSMUST00000207546.2
|
Gm45194
|
predicted gene 45194 |
| chr18_-_88945571 | 0.08 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr6_+_77219627 | 0.07 |
ENSMUST00000159616.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr16_-_89284830 | 0.07 |
ENSMUST00000072280.5
|
Krtap8-1
|
keratin associated protein 8-1 |
| chr1_+_172383499 | 0.07 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr13_-_103042554 | 0.07 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_31417834 | 0.07 |
ENSMUST00000236793.2
|
Tmprss3
|
transmembrane protease, serine 3 |
| chr16_-_48592319 | 0.07 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr11_-_42070517 | 0.07 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr3_-_37286714 | 0.07 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr7_-_6525801 | 0.07 |
ENSMUST00000213504.2
ENSMUST00000216447.2 ENSMUST00000213656.2 ENSMUST00000207820.3 |
Olfr1349
|
olfactory receptor 1349 |
| chr12_-_56581823 | 0.07 |
ENSMUST00000178477.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr19_+_55730242 | 0.06 |
ENSMUST00000111662.11
ENSMUST00000041717.14 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr2_-_86109346 | 0.06 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr6_+_77219698 | 0.06 |
ENSMUST00000161677.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr7_+_23451695 | 0.06 |
ENSMUST00000228331.2
|
Vmn1r174
|
vomeronasal 1 receptor 174 |
| chr19_+_16933471 | 0.06 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chr7_-_86016045 | 0.06 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr12_-_81579614 | 0.06 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr1_+_153541412 | 0.06 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr9_-_99302205 | 0.06 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr3_-_108133914 | 0.06 |
ENSMUST00000141387.4
|
Sypl2
|
synaptophysin-like 2 |
| chr4_+_118577335 | 0.06 |
ENSMUST00000217334.3
ENSMUST00000213436.3 ENSMUST00000216242.3 |
Olfr1340
|
olfactory receptor 1340 |
| chr7_+_92729067 | 0.06 |
ENSMUST00000051179.12
|
Fam181b
|
family with sequence similarity 181, member B |
| chr11_+_73241609 | 0.06 |
ENSMUST00000120137.3
|
Olfr20
|
olfactory receptor 20 |
| chrX_+_152020744 | 0.06 |
ENSMUST00000112574.9
|
Klf8
|
Kruppel-like factor 8 |
| chr19_-_5610628 | 0.06 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr14_-_20718337 | 0.06 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chr2_-_105832353 | 0.05 |
ENSMUST00000155811.2
|
Dnajc24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr4_+_150322151 | 0.05 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chr13_-_21744063 | 0.05 |
ENSMUST00000217519.2
|
Olfr1535
|
olfactory receptor 1535 |
| chr19_+_13890894 | 0.05 |
ENSMUST00000216623.2
ENSMUST00000216835.2 |
Olfr1505
|
olfactory receptor 1505 |
| chr7_-_106531426 | 0.05 |
ENSMUST00000215468.2
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr12_-_112477536 | 0.05 |
ENSMUST00000066791.7
|
Tmem179
|
transmembrane protein 179 |
| chr10_+_50770836 | 0.05 |
ENSMUST00000219436.2
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr13_-_103042294 | 0.05 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_84500854 | 0.05 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr6_-_138403732 | 0.05 |
ENSMUST00000162932.2
|
Lmo3
|
LIM domain only 3 |
| chr6_-_146855880 | 0.05 |
ENSMUST00000111622.2
ENSMUST00000036592.15 |
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene |
| chr7_+_23752679 | 0.05 |
ENSMUST00000236959.2
|
Vmn1r183
|
vomeronasal 1 receptor 183 |
| chr5_+_117979899 | 0.05 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr19_-_55229668 | 0.05 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr17_+_17622934 | 0.05 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr13_+_110063364 | 0.04 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr11_+_67128843 | 0.04 |
ENSMUST00000018632.11
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr1_+_153541339 | 0.04 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr8_-_68270936 | 0.04 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_-_73283010 | 0.04 |
ENSMUST00000151939.2
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr16_+_25620652 | 0.04 |
ENSMUST00000115304.8
ENSMUST00000115305.2 ENSMUST00000040231.13 ENSMUST00000115306.8 |
Trp63
|
transformation related protein 63 |
| chr19_-_41921676 | 0.04 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr2_-_87504008 | 0.04 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr11_-_99996452 | 0.04 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr7_-_126183716 | 0.04 |
ENSMUST00000150311.8
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr3_+_29568055 | 0.04 |
ENSMUST00000140288.2
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr13_-_114595122 | 0.04 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chr11_+_73262072 | 0.04 |
ENSMUST00000078952.9
ENSMUST00000120401.9 ENSMUST00000170592.4 |
Olfr376
|
olfactory receptor 376 |
| chr11_-_99265721 | 0.04 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr11_-_65160767 | 0.04 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr8_-_25215778 | 0.03 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr10_-_70491764 | 0.03 |
ENSMUST00000162144.2
ENSMUST00000162793.8 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr19_+_55730316 | 0.03 |
ENSMUST00000111658.10
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr11_-_65160810 | 0.03 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chr2_+_88505972 | 0.03 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr7_+_126184141 | 0.03 |
ENSMUST00000137646.8
|
Apobr
|
apolipoprotein B receptor |
| chr10_-_129107354 | 0.03 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr4_+_54947976 | 0.03 |
ENSMUST00000098070.10
|
Zfp462
|
zinc finger protein 462 |
| chr19_+_55730488 | 0.03 |
ENSMUST00000111659.9
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr1_-_143652711 | 0.03 |
ENSMUST00000159879.2
|
Ro60
|
Ro60, Y RNA binding protein |
| chr5_+_13449276 | 0.03 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr18_+_20509771 | 0.03 |
ENSMUST00000076737.7
|
Dsg1b
|
desmoglein 1 beta |
| chr12_+_33978401 | 0.02 |
ENSMUST00000061035.4
|
Ferd3l
|
Fer3 like bHLH transcription factor |
| chr4_+_150321659 | 0.02 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr11_-_105346120 | 0.02 |
ENSMUST00000138977.8
|
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr7_-_10292412 | 0.02 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr15_+_35371644 | 0.02 |
ENSMUST00000227455.2
|
Vps13b
|
vacuolar protein sorting 13B |
| chr7_+_23400128 | 0.02 |
ENSMUST00000226233.2
ENSMUST00000227987.2 |
Vmn1r173
|
vomeronasal 1 receptor 173 |
| chr7_-_126183392 | 0.02 |
ENSMUST00000128970.8
ENSMUST00000116269.9 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr4_-_91288221 | 0.02 |
ENSMUST00000102799.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr2_+_86122799 | 0.02 |
ENSMUST00000217166.2
|
Olfr1052
|
olfactory receptor 1052 |
| chr9_-_19289965 | 0.02 |
ENSMUST00000216839.3
|
Olfr847
|
olfactory receptor 847 |
| chr15_-_101441254 | 0.01 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84 |
| chr7_+_23239157 | 0.01 |
ENSMUST00000235361.2
|
Vmn1r168
|
vomeronasal 1 receptor 168 |
| chr4_-_43710231 | 0.01 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr14_-_70043079 | 0.01 |
ENSMUST00000022665.4
|
Rhobtb2
|
Rho-related BTB domain containing 2 |
| chr4_-_133329479 | 0.01 |
ENSMUST00000057311.4
|
Sfn
|
stratifin |
| chr17_-_90395568 | 0.01 |
ENSMUST00000173222.2
|
Nrxn1
|
neurexin I |
| chr10_+_79746690 | 0.01 |
ENSMUST00000181321.2
|
Gm26602
|
predicted gene, 26602 |
| chr1_-_158183894 | 0.01 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr2_-_111820618 | 0.01 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chrX_+_82898974 | 0.01 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 1.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 1.4 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 1.6 | GO:1901750 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.5 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.2 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.5 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 1.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.0 | 0.6 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.1 | GO:1904753 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.0 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.3 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |