Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
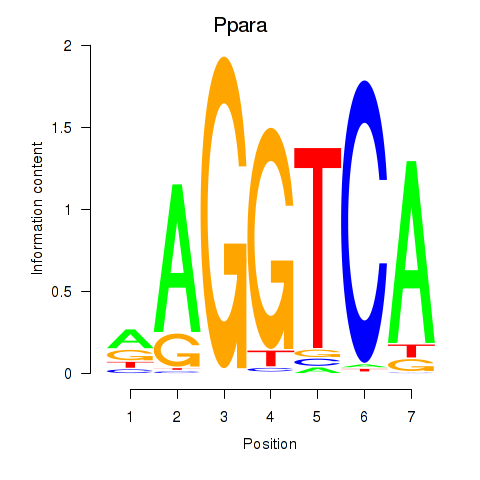
Results for Ppara
Z-value: 1.66
Transcription factors associated with Ppara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppara
|
ENSMUSG00000022383.14 | Ppara |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ppara | mm39_v1_chr15_+_85620308_85620332 | -0.20 | 2.4e-01 | Click! |
Activity profile of Ppara motif
Sorted Z-values of Ppara motif
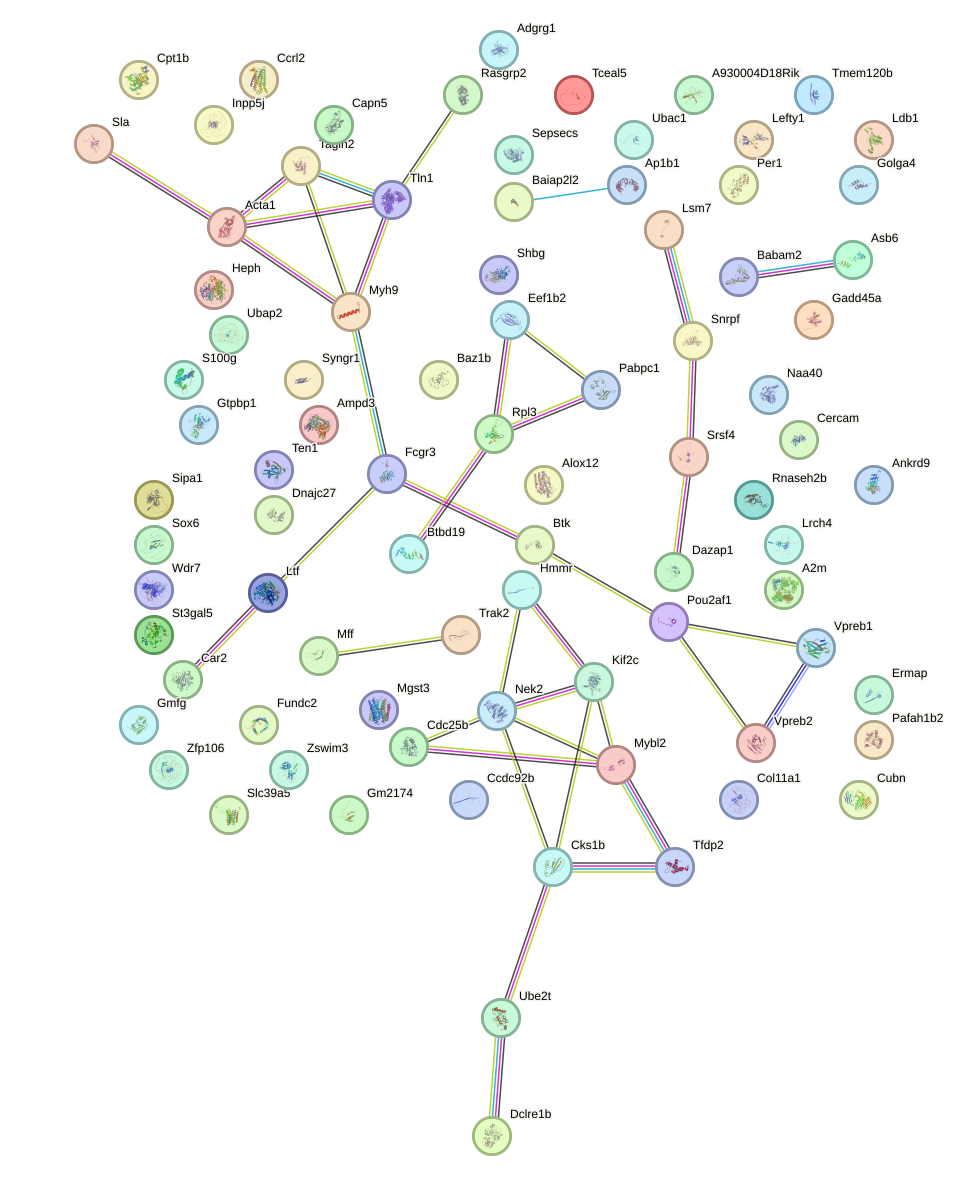
Network of associatons between targets according to the STRING database.
First level regulatory network of Ppara
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_69508706 | 10.28 |
ENSMUST00000005334.3
|
Shbg
|
sex hormone binding globulin |
| chr2_+_131028861 | 6.96 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr1_-_167221344 | 6.83 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr3_+_14951264 | 6.61 |
ENSMUST00000192609.6
|
Car2
|
carbonic anhydrase 2 |
| chr3_+_14951478 | 5.90 |
ENSMUST00000029078.9
|
Car2
|
carbonic anhydrase 2 |
| chr9_+_110848339 | 5.47 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr6_+_121613177 | 5.33 |
ENSMUST00000032203.9
|
A2m
|
alpha-2-macroglobulin |
| chr2_+_162916551 | 5.25 |
ENSMUST00000142729.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr15_-_36598263 | 4.93 |
ENSMUST00000155116.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_+_109425769 | 4.70 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr15_+_79975520 | 4.55 |
ENSMUST00000009728.13
ENSMUST00000009727.12 |
Syngr1
|
synaptogyrin 1 |
| chr7_-_100164007 | 4.47 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr10_-_128237087 | 4.30 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr5_+_135216090 | 4.22 |
ENSMUST00000002825.6
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr4_-_131802606 | 4.19 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr4_-_119047202 | 4.10 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr2_-_131001916 | 4.09 |
ENSMUST00000103188.10
ENSMUST00000133602.8 ENSMUST00000028800.12 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr2_-_25911691 | 4.09 |
ENSMUST00000036509.14
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr11_+_4936824 | 4.02 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr16_-_16687119 | 3.96 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr2_-_25911544 | 3.95 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr8_+_95703728 | 3.87 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr6_-_67014383 | 3.78 |
ENSMUST00000043098.9
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr15_+_73594965 | 3.66 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr9_+_96140781 | 3.62 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr15_+_73595012 | 3.61 |
ENSMUST00000230044.2
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr4_-_119047167 | 3.51 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr8_-_124621483 | 3.51 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chrX_-_135104589 | 3.46 |
ENSMUST00000066819.11
|
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr17_-_71575584 | 3.45 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr2_+_29759495 | 3.43 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr1_+_191553556 | 3.42 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr10_+_80100812 | 3.39 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr18_-_57058581 | 3.37 |
ENSMUST00000102912.8
|
Marchf3
|
membrane associated ring-CH-type finger 3 |
| chr10_-_93425553 | 3.34 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr4_-_43558386 | 3.33 |
ENSMUST00000130353.2
|
Tln1
|
talin 1 |
| chr8_+_85696695 | 3.30 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr11_-_70146156 | 3.25 |
ENSMUST00000108574.3
ENSMUST00000000329.9 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr11_-_3454766 | 3.18 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr11_+_74510413 | 3.12 |
ENSMUST00000100866.3
|
Ccdc92b
|
coiled-coil domain containing 92B |
| chr11_-_40624200 | 3.08 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr9_+_65008735 | 3.07 |
ENSMUST00000213533.2
ENSMUST00000035499.5 ENSMUST00000077696.13 ENSMUST00000166273.2 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr7_-_115630282 | 3.03 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr12_-_110945415 | 3.03 |
ENSMUST00000135131.2
ENSMUST00000043459.13 ENSMUST00000128353.8 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr7_-_97811525 | 2.98 |
ENSMUST00000107112.2
|
Capn5
|
calpain 5 |
| chr14_+_62529924 | 2.94 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr11_+_116089678 | 2.92 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr1_-_59012579 | 2.88 |
ENSMUST00000173590.2
ENSMUST00000027186.12 |
Trak2
|
trafficking protein, kinesin binding 2 |
| chr7_+_110368037 | 2.85 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr4_-_119047180 | 2.84 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047146 | 2.83 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr19_-_7218363 | 2.81 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr11_+_68989763 | 2.80 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr8_+_85696453 | 2.79 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_85696396 | 2.78 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr3_+_113824181 | 2.75 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr1_+_82702598 | 2.72 |
ENSMUST00000078332.13
ENSMUST00000073025.12 ENSMUST00000161648.8 ENSMUST00000160786.8 ENSMUST00000162003.8 |
Mff
|
mitochondrial fission factor |
| chr6_-_67014348 | 2.63 |
ENSMUST00000204369.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chrX_-_149372840 | 2.63 |
ENSMUST00000112725.8
ENSMUST00000112720.8 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr2_-_13496624 | 2.62 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr3_-_103716593 | 2.58 |
ENSMUST00000063502.13
ENSMUST00000106832.2 ENSMUST00000106834.8 ENSMUST00000029435.15 |
Dclre1b
|
DNA cross-link repair 1B |
| chr5_+_137628377 | 2.53 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr6_-_67014191 | 2.51 |
ENSMUST00000204282.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr1_-_167112784 | 2.50 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr5_+_123214332 | 2.50 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr8_+_95722289 | 2.44 |
ENSMUST00000211984.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr16_+_17798292 | 2.43 |
ENSMUST00000075371.5
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr4_-_117035922 | 2.41 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr6_+_72074718 | 2.40 |
ENSMUST00000187007.3
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr2_+_164647002 | 2.36 |
ENSMUST00000052107.5
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr9_+_51124983 | 2.35 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr15_-_89310060 | 2.35 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr7_-_126302315 | 2.35 |
ENSMUST00000173108.8
ENSMUST00000205515.2 |
Coro1a
|
coronin, actin binding protein 1A |
| chr15_+_79578141 | 2.33 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr4_-_131802561 | 2.33 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr7_-_115445352 | 2.31 |
ENSMUST00000206369.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr1_+_172328768 | 2.30 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chrX_+_74425990 | 2.24 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr7_-_110581652 | 2.23 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chrX_+_55493325 | 2.23 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr15_-_79169671 | 2.22 |
ENSMUST00000170955.2
ENSMUST00000165408.8 |
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr15_-_79976016 | 2.22 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr3_-_89325594 | 2.22 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr10_-_80691009 | 2.21 |
ENSMUST00000220225.2
ENSMUST00000035775.9 |
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_+_110367375 | 2.20 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr2_-_30720345 | 2.20 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr15_-_63932288 | 2.20 |
ENSMUST00000063838.11
ENSMUST00000228908.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr9_+_96140750 | 2.16 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr9_+_118335294 | 2.14 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr16_+_43960183 | 2.10 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr15_-_66673425 | 2.10 |
ENSMUST00000168589.8
|
Sla
|
src-like adaptor |
| chrX_-_133483849 | 2.10 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chrX_-_135104386 | 2.08 |
ENSMUST00000151592.8
ENSMUST00000131510.2 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr1_+_172327569 | 2.08 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr4_-_116982804 | 2.03 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr4_+_131600918 | 2.03 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr18_+_63841756 | 2.03 |
ENSMUST00000072726.7
ENSMUST00000235648.2 ENSMUST00000236879.2 |
Wdr7
|
WD repeat domain 7 |
| chr3_+_95071617 | 2.03 |
ENSMUST00000168321.8
ENSMUST00000107217.6 ENSMUST00000202315.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr8_+_95703506 | 2.01 |
ENSMUST00000212581.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_-_110886306 | 2.00 |
ENSMUST00000195968.2
ENSMUST00000111888.3 |
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr19_-_7218512 | 1.98 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr19_-_5713648 | 1.97 |
ENSMUST00000080824.13
ENSMUST00000237874.2 ENSMUST00000071857.13 ENSMUST00000236464.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr9_-_21202545 | 1.96 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chrX_-_161747552 | 1.95 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr12_-_110945376 | 1.95 |
ENSMUST00000142012.2
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr1_+_134890288 | 1.95 |
ENSMUST00000027687.8
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr19_+_37184927 | 1.93 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr4_-_41275091 | 1.92 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr1_+_63215976 | 1.91 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr7_-_115445315 | 1.89 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_167112170 | 1.86 |
ENSMUST00000192269.3
|
Uck2
|
uridine-cytidine kinase 2 |
| chr1_+_63216281 | 1.86 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_-_120394288 | 1.84 |
ENSMUST00000055241.13
ENSMUST00000135625.8 |
Zfp106
|
zinc finger protein 106 |
| chr19_-_46033353 | 1.83 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr1_-_170886924 | 1.83 |
ENSMUST00000164044.8
ENSMUST00000169017.8 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr9_-_45896663 | 1.81 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr15_-_77653979 | 1.81 |
ENSMUST00000229259.2
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr1_+_180762587 | 1.80 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr5_+_31855009 | 1.79 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr19_+_6450553 | 1.79 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr15_-_63932176 | 1.77 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr12_+_4132567 | 1.76 |
ENSMUST00000020986.15
ENSMUST00000049584.6 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr7_+_28140352 | 1.75 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr2_+_153583194 | 1.73 |
ENSMUST00000028981.9
|
Mapre1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr9_+_115738523 | 1.73 |
ENSMUST00000119291.8
ENSMUST00000069651.13 |
Gadl1
|
glutamate decarboxylase-like 1 |
| chr1_+_130659700 | 1.73 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr9_-_45896110 | 1.70 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr10_-_119075910 | 1.70 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr9_-_21202693 | 1.70 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr11_-_96868483 | 1.69 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr7_-_24705320 | 1.66 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr19_-_5713701 | 1.64 |
ENSMUST00000164304.9
ENSMUST00000237544.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr11_+_97340962 | 1.63 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr16_-_19801781 | 1.61 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr12_-_110945052 | 1.61 |
ENSMUST00000140788.8
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr2_+_155453103 | 1.60 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr15_-_98505508 | 1.60 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr7_+_24161899 | 1.60 |
ENSMUST00000002284.11
|
Plaur
|
plasminogen activator, urokinase receptor |
| chr5_-_5744024 | 1.60 |
ENSMUST00000115425.9
ENSMUST00000115427.8 ENSMUST00000115424.9 ENSMUST00000015797.11 |
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr6_+_125122172 | 1.60 |
ENSMUST00000119527.8
ENSMUST00000117675.8 |
Iffo1
|
intermediate filament family orphan 1 |
| chr17_+_69463786 | 1.58 |
ENSMUST00000112680.8
ENSMUST00000080208.7 ENSMUST00000225977.2 |
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr6_+_113508636 | 1.58 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr4_+_33310306 | 1.57 |
ENSMUST00000108153.9
ENSMUST00000029942.8 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chrX_-_52357897 | 1.56 |
ENSMUST00000114838.8
|
Fam122b
|
family with sequence similarity 122, member B |
| chr8_+_71921824 | 1.56 |
ENSMUST00000124745.8
ENSMUST00000138892.2 ENSMUST00000147642.2 |
Dda1
|
DET1 and DDB1 associated 1 |
| chr19_+_8568618 | 1.55 |
ENSMUST00000170817.2
ENSMUST00000010251.11 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr6_+_123099615 | 1.55 |
ENSMUST00000161636.8
ENSMUST00000161365.8 |
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr13_+_108350923 | 1.54 |
ENSMUST00000022207.10
|
Elovl7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| chr4_-_116228921 | 1.54 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr7_+_79836581 | 1.54 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr8_-_105350898 | 1.53 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr4_-_129436465 | 1.53 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr5_-_110987604 | 1.52 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr9_+_98178608 | 1.52 |
ENSMUST00000112935.8
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_+_129435448 | 1.51 |
ENSMUST00000049262.14
ENSMUST00000163034.8 ENSMUST00000160276.2 |
Sirpa
|
signal-regulatory protein alpha |
| chr8_-_4309257 | 1.51 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr16_-_4340920 | 1.49 |
ENSMUST00000090500.10
ENSMUST00000023161.8 |
Srl
|
sarcalumenin |
| chr12_-_84240781 | 1.48 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr12_-_40298072 | 1.48 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr11_+_31822211 | 1.48 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr9_-_42372710 | 1.46 |
ENSMUST00000066179.14
|
Tbcel
|
tubulin folding cofactor E-like |
| chr5_-_31854942 | 1.46 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr19_-_37184692 | 1.46 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr8_-_81466126 | 1.45 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr4_-_126096376 | 1.45 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr7_+_130467564 | 1.45 |
ENSMUST00000075181.11
ENSMUST00000151119.9 ENSMUST00000048180.12 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr6_+_85408953 | 1.45 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr13_+_52750883 | 1.44 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr5_+_110987839 | 1.40 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr17_+_84013575 | 1.40 |
ENSMUST00000112350.8
ENSMUST00000112349.9 ENSMUST00000112352.10 ENSMUST00000067826.15 |
Mta3
|
metastasis associated 3 |
| chr7_+_30450896 | 1.39 |
ENSMUST00000182229.8
ENSMUST00000080518.14 ENSMUST00000182227.8 ENSMUST00000182721.8 |
Sbsn
|
suprabasin |
| chr15_+_61859255 | 1.39 |
ENSMUST00000160009.2
|
Myc
|
myelocytomatosis oncogene |
| chr7_-_19449319 | 1.38 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr1_+_75377616 | 1.37 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr1_+_172327812 | 1.37 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr17_-_24360516 | 1.36 |
ENSMUST00000115411.8
ENSMUST00000115409.9 ENSMUST00000115407.9 ENSMUST00000102927.10 |
Pdpk1
|
3-phosphoinositide dependent protein kinase 1 |
| chr1_+_12788720 | 1.36 |
ENSMUST00000088585.10
|
Sulf1
|
sulfatase 1 |
| chr6_-_76474767 | 1.36 |
ENSMUST00000097218.7
|
Gm9008
|
predicted pseudogene 9008 |
| chr19_+_47079187 | 1.36 |
ENSMUST00000072141.4
|
Pdcd11
|
programmed cell death 11 |
| chr7_+_141995545 | 1.36 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr9_+_65494469 | 1.35 |
ENSMUST00000239405.2
ENSMUST00000047099.13 ENSMUST00000131483.3 ENSMUST00000141046.3 |
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chr6_+_120813199 | 1.35 |
ENSMUST00000009256.4
|
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr14_+_79753055 | 1.34 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr1_-_84817022 | 1.33 |
ENSMUST00000189496.7
ENSMUST00000027421.13 ENSMUST00000186894.7 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chrX_-_12026594 | 1.33 |
ENSMUST00000043441.13
|
Bcor
|
BCL6 interacting corepressor |
| chr11_-_61384998 | 1.33 |
ENSMUST00000101085.9
ENSMUST00000079080.13 ENSMUST00000108714.2 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr7_-_44888220 | 1.31 |
ENSMUST00000210372.2
ENSMUST00000209779.2 ENSMUST00000098461.10 ENSMUST00000211373.2 |
Cd37
|
CD37 antigen |
| chr1_-_152262425 | 1.31 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr2_+_75662511 | 1.31 |
ENSMUST00000047232.14
ENSMUST00000111952.9 |
Agps
|
alkylglycerone phosphate synthase |
| chr15_+_84553801 | 1.30 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr19_+_4644425 | 1.29 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr10_+_58207229 | 1.29 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chrX_+_70600481 | 1.29 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr4_-_133981387 | 1.28 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr6_-_83030759 | 1.28 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr14_+_75373915 | 1.27 |
ENSMUST00000122840.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chrX_-_48877080 | 1.27 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chrX_-_56384089 | 1.27 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr12_-_69205882 | 1.27 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr7_-_19410749 | 1.27 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.5 | GO:2000878 | positive regulation of cellular pH reduction(GO:0032849) dipeptide transmembrane transport(GO:0035442) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) positive regulation of oligopeptide transport(GO:2000878) positive regulation of dipeptide transport(GO:2000880) regulation of dipeptide transmembrane transport(GO:2001148) positive regulation of dipeptide transmembrane transport(GO:2001150) |
| 1.8 | 5.3 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.5 | 4.4 | GO:0032262 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 1.4 | 5.5 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.1 | 4.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.9 | 2.8 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.9 | 3.5 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 0.9 | 4.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.9 | 7.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.7 | 8.9 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.7 | 2.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.7 | 3.6 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.7 | 4.9 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.7 | 3.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.7 | 7.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.6 | 3.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.6 | 2.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.6 | 8.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 1.8 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.6 | 1.8 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.6 | 1.8 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.6 | 1.8 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.6 | 2.3 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.5 | 1.6 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.5 | 4.2 | GO:0032796 | uropod organization(GO:0032796) |
| 0.5 | 1.5 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.5 | 5.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 1.5 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.5 | 1.4 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.5 | 2.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.5 | 6.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.5 | 2.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.5 | 1.4 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.4 | 1.3 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.4 | 2.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.4 | 1.7 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 2.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 1.2 | GO:0061723 | glycophagy(GO:0061723) |
| 0.4 | 1.2 | GO:0051695 | actin filament uncapping(GO:0051695) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.4 | 2.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 7.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.4 | 2.8 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 3.9 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 2.7 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.4 | 1.9 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.4 | 1.6 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.4 | 1.2 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.4 | 7.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.4 | 1.1 | GO:0055130 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 1.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.4 | 1.5 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.4 | 1.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 1.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.4 | 3.5 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.4 | 3.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.4 | 3.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 | 1.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.3 | 1.4 | GO:0090095 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.3 | 2.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 1.0 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.3 | 8.8 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.3 | 1.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 1.9 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.3 | 1.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.3 | 7.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 0.6 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.3 | 1.8 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.3 | 1.2 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.3 | 1.2 | GO:2000705 | positive regulation of relaxation of muscle(GO:1901079) regulation of dense core granule biogenesis(GO:2000705) |
| 0.3 | 1.1 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.3 | 0.8 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.3 | 1.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.3 | 1.0 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 | 2.8 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.3 | 0.5 | GO:2000739 | regulation of mesenchymal stem cell differentiation(GO:2000739) |
| 0.3 | 0.8 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.3 | 1.5 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.3 | 2.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 0.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 1.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 0.5 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.2 | 2.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 0.7 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 3.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.7 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.2 | 0.9 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 1.2 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.2 | 5.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 2.5 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.2 | 0.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 1.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 1.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 1.3 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.2 | 0.9 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 | 1.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 0.9 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.2 | 0.8 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 5.5 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 6.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 0.6 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 1.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.7 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.6 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 4.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 2.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.9 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.2 | 1.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.7 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 1.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.7 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 1.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 1.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 0.7 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.2 | 1.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 1.5 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.2 | 4.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.0 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 1.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 0.5 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 0.5 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 1.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.2 | 0.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 2.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.9 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.3 | GO:0070428 | interleukin-13 biosynthetic process(GO:0042231) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 2.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.6 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.4 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 1.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 2.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 1.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.5 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 0.7 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.4 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.1 | 0.5 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 5.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.3 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 2.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.7 | GO:0048298 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.5 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 1.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.4 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.1 | 0.7 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 1.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 6.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 4.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.6 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.1 | 0.5 | GO:1903936 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 2.0 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.4 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 1.5 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 1.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.2 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 2.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 2.5 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 1.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.4 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 1.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.8 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 2.3 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.4 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.3 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 2.8 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 0.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 5.9 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 0.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.3 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 3.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 1.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.1 | 1.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.1 | 3.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.9 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 2.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.6 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 0.2 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 4.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.2 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.4 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.2 | GO:0035037 | sperm entry(GO:0035037) |
| 0.1 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 2.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 1.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 2.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.3 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 0.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.9 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 1.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.4 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 1.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 2.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 4.0 | GO:1904377 | positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.1 | 1.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.3 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.4 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 1.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.5 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.1 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.5 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 2.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.1 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.4 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 0.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0002551 | mast cell chemotaxis(GO:0002551) |
| 0.1 | 1.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 2.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.8 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 2.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.4 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 1.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 1.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.4 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 4.6 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.7 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 3.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 1.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 2.4 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 2.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 2.1 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 9.5 | GO:0048588 | developmental cell growth(GO:0048588) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 2.0 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.5 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.5 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.5 | GO:0071549 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.6 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.4 | GO:0035459 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 1.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 2.2 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 1.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.3 | GO:2000679 | positive regulation of transcription regulatory region DNA binding(GO:2000679) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.8 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.0 | 0.9 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.8 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.3 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 1.1 | GO:0031960 | response to corticosteroid(GO:0031960) |
| 0.0 | 3.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.0 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.0 | 0.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 3.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 1.2 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 1.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.8 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.8 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.2 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.4 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.0 | GO:0060744 | positive regulation of exit from mitosis(GO:0031536) negative regulation of mammary gland epithelial cell proliferation(GO:0033600) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.0 | 0.6 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.5 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:1990879 | CST complex(GO:1990879) |
| 0.9 | 5.5 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.9 | 5.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.8 | 8.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.6 | 3.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.5 | 3.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 2.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.5 | 3.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 3.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.4 | 2.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 1.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.4 | 3.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.4 | 3.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 2.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 8.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 2.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 0.9 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.3 | 0.9 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 1.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 0.9 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 3.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 2.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 2.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 2.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 2.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 2.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 2.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.7 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 2.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 2.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 2.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 1.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 0.7 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.2 | 5.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 2.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.9 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.6 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 1.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 7.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.4 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 0.7 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 2.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.5 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.1 | 0.4 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 6.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 4.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 4.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 4.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 2.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.7 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 2.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 1.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 7.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 3.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 2.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 11.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.3 | GO:0005608 | laminin-1 complex(GO:0005606) laminin-3 complex(GO:0005608) |
| 0.1 | 20.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.4 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.6 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 4.0 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 2.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 3.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 2.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 12.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 4.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 4.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 3.2 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 4.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 7.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 7.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.9 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 1.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 13.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 2.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.9 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 10.5 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 8.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 3.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 1.1 | 11.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.1 | 3.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.8 | 3.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.8 | 2.4 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.7 | 2.9 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.6 | 1.9 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.6 | 9.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 4.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.6 | 4.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.6 | 1.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.6 | 2.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.6 | 2.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.5 | 4.8 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.5 | 1.6 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 7.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 1.5 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.5 | 5.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 3.9 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 2.0 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.5 | 1.8 | GO:0038100 | nodal binding(GO:0038100) |
| 0.4 | 5.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 3.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 2.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.4 | 2.7 | GO:0002135 | CTP binding(GO:0002135) |
| 0.4 | 2.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 1.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 2.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 2.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 1.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 0.9 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.3 | 3.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 0.8 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.3 | 1.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.3 | 1.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 6.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 3.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.3 | 1.1 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 1.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 1.5 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 1.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.2 | 2.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 1.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 2.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 5.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 6.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 4.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 5.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.2 | 1.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 0.7 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.2 | 0.9 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 3.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.6 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.5 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.2 | 0.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 5.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 5.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.2 | 3.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 1.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 1.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 2.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.9 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.2 | 3.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.9 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.2 | 0.6 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.1 | 0.4 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 0.9 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 1.0 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 5.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.4 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.1 | 1.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 1.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 5.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.5 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 0.5 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 2.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 2.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.7 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 2.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.7 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.0 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 1.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 2.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 14.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.3 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 6.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 3.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 5.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 3.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 3.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.6 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 2.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.0 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 1.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 3.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 4.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 3.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 3.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 5.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 1.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 2.1 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 3.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 4.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 4.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 2.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 2.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 2.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 6.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 3.5 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 8.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 4.5 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 3.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0017002 | activin receptor activity, type I(GO:0016361) activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.6 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 5.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 10.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 12.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 9.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 7.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 8.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 14.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 8.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 5.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 5.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 3.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 4.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 2.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 8.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 4.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 3.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 4.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 5.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 1.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 5.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 1.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 6.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 5.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 2.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 1.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 5.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 3.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 3.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 5.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 3.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 5.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.1 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 5.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 4.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 5.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 3.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.7 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.8 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |