Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
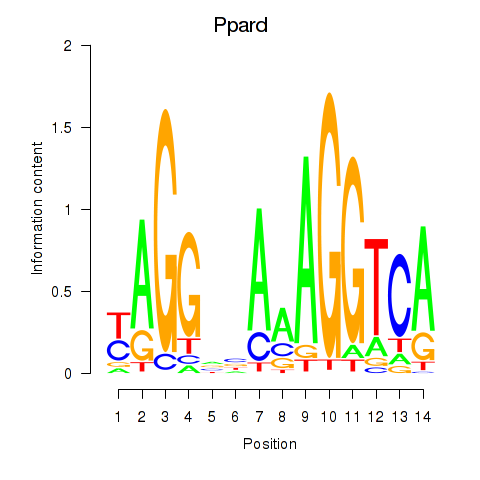
Results for Ppard
Z-value: 0.98
Transcription factors associated with Ppard
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppard
|
ENSMUSG00000002250.17 | Ppard |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ppard | mm39_v1_chr17_+_28491085_28491135 | -0.14 | 4.1e-01 | Click! |
Activity profile of Ppard motif
Sorted Z-values of Ppard motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ppard
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_46146558 | 4.94 |
ENSMUST00000121916.8
ENSMUST00000034586.9 |
Apoc3
|
apolipoprotein C-III |
| chr3_-_79535966 | 3.77 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr16_+_22769822 | 3.71 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_22769844 | 3.50 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr9_-_46146928 | 3.23 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr9_+_108539296 | 2.50 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr3_-_79536166 | 2.40 |
ENSMUST00000029386.14
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr3_+_98187743 | 2.33 |
ENSMUST00000120541.8
ENSMUST00000090746.3 |
Hmgcs2
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
| chr15_-_89310060 | 1.90 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr2_+_92205651 | 1.84 |
ENSMUST00000028650.9
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr4_+_59581617 | 1.70 |
ENSMUST00000107528.8
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr1_+_139429430 | 1.61 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr15_-_102097466 | 1.48 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr15_-_102097387 | 1.42 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr11_-_69906171 | 1.30 |
ENSMUST00000018718.8
ENSMUST00000102574.10 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr17_-_46749370 | 1.28 |
ENSMUST00000087012.7
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr11_-_73217298 | 1.26 |
ENSMUST00000155630.9
|
Aspa
|
aspartoacylase |
| chr11_-_83483807 | 1.18 |
ENSMUST00000019071.4
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr10_-_125164399 | 1.16 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr10_+_3490232 | 1.13 |
ENSMUST00000019896.5
|
Iyd
|
iodotyrosine deiodinase |
| chr5_+_114284585 | 1.08 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr3_-_137837117 | 1.06 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr7_+_43093507 | 1.04 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr10_-_125164826 | 1.02 |
ENSMUST00000211781.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr11_-_73217633 | 1.00 |
ENSMUST00000134079.2
|
Aspa
|
aspartoacylase |
| chr1_-_120047868 | 0.88 |
ENSMUST00000112648.8
|
Dbi
|
diazepam binding inhibitor |
| chr2_-_103315483 | 0.79 |
ENSMUST00000028610.10
|
Cat
|
catalase |
| chr2_-_103315418 | 0.76 |
ENSMUST00000111168.4
|
Cat
|
catalase |
| chr18_+_74912268 | 0.73 |
ENSMUST00000041053.11
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
| chr19_-_6899173 | 0.69 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr5_+_36622342 | 0.67 |
ENSMUST00000031099.4
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr17_-_46749320 | 0.66 |
ENSMUST00000233575.2
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr2_+_162829250 | 0.64 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr16_-_84632383 | 0.61 |
ENSMUST00000114191.8
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr16_-_23339329 | 0.60 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr2_+_162829422 | 0.58 |
ENSMUST00000117123.2
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_+_171052623 | 0.58 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr9_+_21914334 | 0.56 |
ENSMUST00000115331.10
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr9_+_21914083 | 0.56 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr1_+_55127110 | 0.55 |
ENSMUST00000075242.7
|
Hspe1
|
heat shock protein 1 (chaperonin 10) |
| chr19_-_6899121 | 0.52 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr10_+_93983844 | 0.52 |
ENSMUST00000105290.9
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr6_-_97436223 | 0.52 |
ENSMUST00000113359.8
|
Frmd4b
|
FERM domain containing 4B |
| chr4_+_148686985 | 0.51 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr2_-_32594156 | 0.50 |
ENSMUST00000127812.3
|
Fpgs
|
folylpolyglutamyl synthetase |
| chrX_+_10118544 | 0.49 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr16_-_84632439 | 0.48 |
ENSMUST00000138279.2
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr9_+_21914296 | 0.48 |
ENSMUST00000003493.9
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr9_+_45924105 | 0.46 |
ENSMUST00000126865.8
|
Sik3
|
SIK family kinase 3 |
| chr17_-_34247016 | 0.40 |
ENSMUST00000236627.2
ENSMUST00000237759.2 ENSMUST00000045467.14 ENSMUST00000114303.4 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chrX_+_10118600 | 0.39 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr16_-_84632513 | 0.39 |
ENSMUST00000023608.14
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr19_+_5510636 | 0.39 |
ENSMUST00000225141.2
ENSMUST00000025847.7 |
Fibp
|
fibroblast growth factor (acidic) intracellular binding protein |
| chr4_+_98434930 | 0.38 |
ENSMUST00000102792.10
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr17_+_34879431 | 0.38 |
ENSMUST00000238967.2
|
Tnxb
|
tenascin XB |
| chr3_+_96737385 | 0.32 |
ENSMUST00000058865.14
|
Pdzk1
|
PDZ domain containing 1 |
| chr1_-_55127183 | 0.31 |
ENSMUST00000027123.15
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr1_-_55127312 | 0.31 |
ENSMUST00000127861.8
ENSMUST00000144077.3 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr2_+_165834546 | 0.31 |
ENSMUST00000109252.8
ENSMUST00000088095.6 |
Ncoa3
|
nuclear receptor coactivator 3 |
| chr18_-_36859732 | 0.30 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr2_-_73741664 | 0.29 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr9_+_21914513 | 0.29 |
ENSMUST00000215795.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr9_+_45924120 | 0.28 |
ENSMUST00000120463.9
ENSMUST00000120247.8 |
Sik3
|
SIK family kinase 3 |
| chr8_+_117822593 | 0.27 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr9_-_108888779 | 0.27 |
ENSMUST00000061973.5
|
Trex1
|
three prime repair exonuclease 1 |
| chr11_-_4654303 | 0.23 |
ENSMUST00000058407.6
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr4_+_98435155 | 0.23 |
ENSMUST00000030290.8
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr11_-_94390788 | 0.21 |
ENSMUST00000127305.2
|
Epn3
|
epsin 3 |
| chr4_-_41048124 | 0.20 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr15_-_50753792 | 0.17 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr19_+_4644365 | 0.16 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr6_-_142418801 | 0.16 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr15_-_89309998 | 0.15 |
ENSMUST00000168376.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr3_+_89958940 | 0.15 |
ENSMUST00000159064.8
|
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr2_+_84891281 | 0.14 |
ENSMUST00000238769.2
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr14_+_14210932 | 0.14 |
ENSMUST00000022271.14
|
Acox2
|
acyl-Coenzyme A oxidase 2, branched chain |
| chr2_+_160730019 | 0.13 |
ENSMUST00000109455.9
ENSMUST00000040872.13 |
Lpin3
|
lipin 3 |
| chr2_-_32594043 | 0.13 |
ENSMUST00000143743.2
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr9_-_21913833 | 0.12 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr1_+_180553569 | 0.11 |
ENSMUST00000027780.6
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr12_+_84498196 | 0.11 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr16_-_91728162 | 0.10 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr9_+_107217786 | 0.10 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr15_+_101184488 | 0.09 |
ENSMUST00000229525.2
ENSMUST00000230525.2 |
Atg101
|
autophagy related 101 |
| chr1_-_133617824 | 0.08 |
ENSMUST00000189524.2
ENSMUST00000169295.8 |
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr1_+_171216480 | 0.07 |
ENSMUST00000056449.9
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr11_+_98337655 | 0.07 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr2_+_90865958 | 0.06 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr15_-_27630724 | 0.06 |
ENSMUST00000059662.8
|
Otulin
|
OTU deubiquitinase with linear linkage specificity |
| chr9_-_21913896 | 0.05 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr12_-_40273173 | 0.03 |
ENSMUST00000001672.12
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr1_+_171386752 | 0.00 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr10_-_14581203 | 0.00 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0097037 | heme export(GO:0097037) |
| 2.0 | 8.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.6 | 2.9 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.4 | 8.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 1.1 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 2.3 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 2.8 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 2.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.3 | 1.8 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.3 | 0.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.2 | 0.6 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.7 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 | 0.9 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.6 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 1.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.5 | GO:0009650 | UV protection(GO:0009650) |
| 0.1 | 0.3 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 1.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 2.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 1.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 | 0.3 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 2.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 1.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0017062 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.7 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.8 | 7.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.7 | 8.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 1.9 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 1.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 1.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 6.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.2 | 6.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.8 | 2.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.8 | 2.3 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.6 | 2.5 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.4 | 1.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.4 | 2.9 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.4 | 1.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 2.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 2.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 0.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 0.9 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.6 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.1 | 0.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 6.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.4 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 1.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.7 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 1.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 1.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 1.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 7.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 5.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 6.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 7.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 2.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |