Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Rara
Z-value: 0.70
Transcription factors associated with Rara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rara
|
ENSMUSG00000037992.17 | Rara |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rara | mm39_v1_chr11_+_98828495_98828559 | -0.12 | 4.8e-01 | Click! |
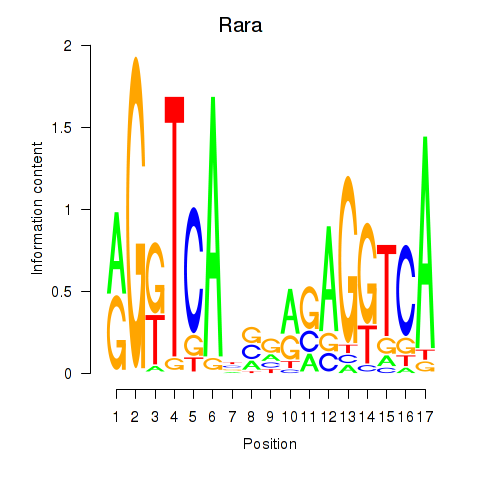
Activity profile of Rara motif
Sorted Z-values of Rara motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rara
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_5865124 | 3.10 |
ENSMUST00000109823.9
ENSMUST00000109822.8 |
Gck
|
glucokinase |
| chr19_+_37686240 | 2.56 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr8_-_72966663 | 2.30 |
ENSMUST00000098630.5
|
Cib3
|
calcium and integrin binding family member 3 |
| chr4_-_46991842 | 2.01 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr19_-_7780025 | 1.86 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr19_-_7779943 | 1.67 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chrX_-_74918122 | 1.44 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr2_+_32498997 | 1.11 |
ENSMUST00000143625.2
ENSMUST00000128811.2 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr6_-_124519240 | 1.00 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr3_+_122305819 | 0.94 |
ENSMUST00000199344.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr5_-_105387395 | 0.90 |
ENSMUST00000065588.7
|
Gbp10
|
guanylate-binding protein 10 |
| chr7_-_44320244 | 0.90 |
ENSMUST00000048102.15
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr12_+_108859557 | 0.90 |
ENSMUST00000221377.2
|
Wdr25
|
WD repeat domain 25 |
| chr10_+_4561974 | 0.88 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr11_+_98851238 | 0.82 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha |
| chr1_-_14374794 | 0.66 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_+_157286124 | 0.64 |
ENSMUST00000193791.6
ENSMUST00000046743.11 ENSMUST00000119891.7 |
Cryzl2
|
crystallin zeta like 2 |
| chr1_-_14374842 | 0.64 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_+_79792238 | 0.49 |
ENSMUST00000135021.2
|
Gask1b
|
golgi associated kinase 1B |
| chr5_-_113285852 | 0.49 |
ENSMUST00000212276.2
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chr16_-_94657531 | 0.48 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr5_-_114411851 | 0.47 |
ENSMUST00000044790.12
|
Foxn4
|
forkhead box N4 |
| chr2_+_120807498 | 0.46 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr2_+_32498716 | 0.44 |
ENSMUST00000129165.2
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr5_+_88073483 | 0.43 |
ENSMUST00000113271.3
|
Csn3
|
casein kappa |
| chr11_-_5211558 | 0.38 |
ENSMUST00000020662.15
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr7_+_16884194 | 0.37 |
ENSMUST00000108491.8
|
Ceacam3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr5_+_114991722 | 0.37 |
ENSMUST00000031547.12
|
4930519G04Rik
|
RIKEN cDNA 4930519G04 gene |
| chr3_-_36529753 | 0.37 |
ENSMUST00000199478.2
|
Anxa5
|
annexin A5 |
| chr7_-_18350423 | 0.36 |
ENSMUST00000057810.7
|
Psg23
|
pregnancy-specific glycoprotein 23 |
| chr4_-_123458465 | 0.35 |
ENSMUST00000238731.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr12_-_54909568 | 0.34 |
ENSMUST00000078124.8
|
Cfl2
|
cofilin 2, muscle |
| chr5_+_88073438 | 0.33 |
ENSMUST00000001667.13
ENSMUST00000113267.8 |
Csn3
|
casein kappa |
| chr4_+_48045143 | 0.33 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr2_+_169475436 | 0.32 |
ENSMUST00000109157.2
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr4_+_150938376 | 0.31 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr4_-_152402915 | 0.30 |
ENSMUST00000170820.3
ENSMUST00000076183.12 |
Rnf207
|
ring finger protein 207 |
| chr8_+_106877025 | 0.29 |
ENSMUST00000212963.2
ENSMUST00000034377.8 |
Pla2g15
|
phospholipase A2, group XV |
| chr1_-_172034251 | 0.28 |
ENSMUST00000155109.2
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr10_-_8638215 | 0.28 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr3_-_88194517 | 0.28 |
ENSMUST00000165196.8
|
Gm38392
|
predicted gene, 38392 |
| chr5_+_14075281 | 0.28 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr6_-_13677928 | 0.27 |
ENSMUST00000203078.2
ENSMUST00000045235.8 |
Bmt2
|
base methyltransferase of 25S rRNA 2 |
| chr3_-_87965757 | 0.26 |
ENSMUST00000029708.8
|
Naxe
|
NAD(P)HX epimerase |
| chrX_-_104918911 | 0.25 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr9_-_56325344 | 0.24 |
ENSMUST00000061552.15
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr17_-_67939702 | 0.24 |
ENSMUST00000097290.4
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr2_+_173561208 | 0.23 |
ENSMUST00000073081.6
|
1700010B08Rik
|
RIKEN cDNA 1700010B08 gene |
| chr7_-_30156826 | 0.23 |
ENSMUST00000045817.14
|
Kirrel2
|
kirre like nephrin family adhesion molecule 2 |
| chr2_+_181007177 | 0.23 |
ENSMUST00000108807.9
|
Zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr3_+_121220146 | 0.23 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr6_+_41107047 | 0.23 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr9_-_56325307 | 0.23 |
ENSMUST00000186735.2
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr4_-_128503774 | 0.23 |
ENSMUST00000141040.2
ENSMUST00000147876.2 ENSMUST00000097877.9 |
Zscan20
|
zinc finger and SCAN domains 20 |
| chr14_+_58308004 | 0.21 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr2_-_5017526 | 0.21 |
ENSMUST00000027980.8
|
Mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr11_+_49184124 | 0.19 |
ENSMUST00000189851.3
|
Olfr1392
|
olfactory receptor 1392 |
| chr6_+_114108190 | 0.18 |
ENSMUST00000032451.9
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
| chrX_-_104919201 | 0.18 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr13_+_83720484 | 0.17 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr17_-_56440817 | 0.17 |
ENSMUST00000167545.3
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr4_+_119590971 | 0.17 |
ENSMUST00000084306.5
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr3_+_79791798 | 0.16 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr14_-_30075424 | 0.16 |
ENSMUST00000224198.3
ENSMUST00000238675.2 ENSMUST00000112249.10 ENSMUST00000224785.3 |
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr8_+_91681550 | 0.16 |
ENSMUST00000210947.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr1_+_93096316 | 0.15 |
ENSMUST00000138595.3
|
Crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr2_-_90900525 | 0.14 |
ENSMUST00000153367.2
ENSMUST00000079976.10 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr14_+_5894220 | 0.13 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr13_+_83720457 | 0.13 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr12_-_108859123 | 0.13 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr3_+_93376058 | 0.13 |
ENSMUST00000029516.3
|
Tchhl1
|
trichohyalin-like 1 |
| chrX_-_58612709 | 0.12 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr14_-_75185281 | 0.12 |
ENSMUST00000088970.7
ENSMUST00000228252.2 |
Lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr14_-_70445086 | 0.12 |
ENSMUST00000022682.6
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr3_+_89136353 | 0.11 |
ENSMUST00000041142.4
|
Muc1
|
mucin 1, transmembrane |
| chr3_+_96011810 | 0.11 |
ENSMUST00000132980.8
ENSMUST00000138206.8 ENSMUST00000090785.9 ENSMUST00000035519.12 |
Otud7b
|
OTU domain containing 7B |
| chr1_+_131678223 | 0.11 |
ENSMUST00000147800.2
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr7_+_15280907 | 0.11 |
ENSMUST00000067288.15
|
Obox1
|
oocyte specific homeobox 1 |
| chr11_-_50844572 | 0.10 |
ENSMUST00000162420.2
ENSMUST00000051159.3 |
Prop1
|
paired like homeodomain factor 1 |
| chr7_-_141009346 | 0.10 |
ENSMUST00000124444.2
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr19_-_12209960 | 0.10 |
ENSMUST00000207710.3
|
Olfr1432
|
olfactory receptor 1432 |
| chrX_+_159865500 | 0.09 |
ENSMUST00000238603.2
|
Scml2
|
Scm polycomb group protein like 2 |
| chr13_-_65200204 | 0.09 |
ENSMUST00000222769.2
|
Prss47
|
protease, serine 47 |
| chr4_+_32238950 | 0.09 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr14_-_70043079 | 0.09 |
ENSMUST00000022665.4
|
Rhobtb2
|
Rho-related BTB domain containing 2 |
| chr7_-_141009264 | 0.08 |
ENSMUST00000164387.2
ENSMUST00000137488.2 ENSMUST00000084436.10 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr8_-_120505104 | 0.08 |
ENSMUST00000212534.2
ENSMUST00000049156.7 |
Meak7
|
MTOR associated protein , eak-7 homolog |
| chr6_-_54949587 | 0.08 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr13_-_65200574 | 0.08 |
ENSMUST00000203968.3
|
Prss47
|
protease, serine 47 |
| chr7_+_4925781 | 0.07 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr1_-_172034354 | 0.07 |
ENSMUST00000013842.12
ENSMUST00000111247.8 |
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr9_-_96513529 | 0.06 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr7_-_18390645 | 0.06 |
ENSMUST00000094793.12
ENSMUST00000182128.2 |
Psg21
|
pregnancy-specific glycoprotein 21 |
| chr3_+_79252668 | 0.06 |
ENSMUST00000164216.6
ENSMUST00000199658.2 |
Gm17359
|
predicted gene, 17359 |
| chr7_+_120234399 | 0.06 |
ENSMUST00000033176.7
ENSMUST00000208400.2 |
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr6_+_67873135 | 0.05 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr5_-_137623331 | 0.05 |
ENSMUST00000031732.14
|
Fbxo24
|
F-box protein 24 |
| chr7_+_17447163 | 0.04 |
ENSMUST00000081907.8
|
Ceacam5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr13_+_95012107 | 0.04 |
ENSMUST00000022195.13
|
Otp
|
orthopedia homeobox |
| chr5_+_53424471 | 0.03 |
ENSMUST00000147148.5
|
Smim20
|
small integral membrane protein 20 |
| chr18_-_39652468 | 0.03 |
ENSMUST00000237944.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr13_+_49574694 | 0.03 |
ENSMUST00000220447.2
|
Ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr4_+_42969934 | 0.03 |
ENSMUST00000190902.2
ENSMUST00000030163.12 |
1700022I11Rik
|
RIKEN cDNA 1700022I11 gene |
| chr15_-_102274781 | 0.02 |
ENSMUST00000078508.7
|
Sp7
|
Sp7 transcription factor 7 |
| chr11_+_87457544 | 0.02 |
ENSMUST00000060360.7
|
Septin4
|
septin 4 |
| chr1_-_163231196 | 0.01 |
ENSMUST00000045138.6
|
Gorab
|
golgin, RAB6-interacting |
| chr7_+_45522551 | 0.01 |
ENSMUST00000211234.2
|
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr3_-_94922525 | 0.00 |
ENSMUST00000128438.2
ENSMUST00000149747.2 ENSMUST00000019482.8 |
Zfp687
|
zinc finger protein 687 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.4 | 2.6 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.3 | 0.8 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 3.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.9 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 1.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.3 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.3 | GO:1900623 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:1901582 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.3 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0021941 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.0 | 2.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.3 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.9 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.1 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 0.1 | GO:0060126 | hypophysis morphogenesis(GO:0048850) somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.8 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 3.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.4 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 2.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 3.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.3 | 0.9 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 3.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.3 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |