Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Rarg
Z-value: 1.15
Transcription factors associated with Rarg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarg
|
ENSMUSG00000001288.16 | Rarg |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarg | mm39_v1_chr15_-_102154874_102154945 | 0.58 | 2.4e-04 | Click! |
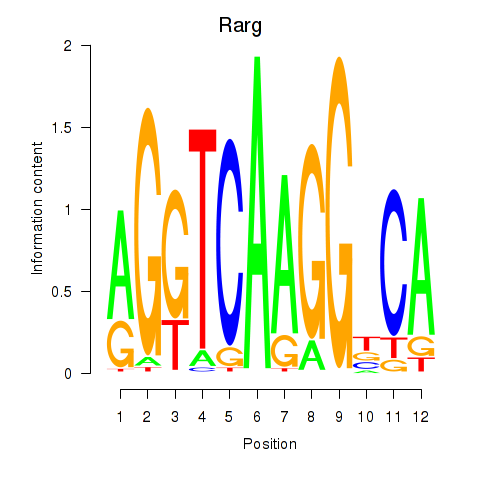
Activity profile of Rarg motif
Sorted Z-values of Rarg motif
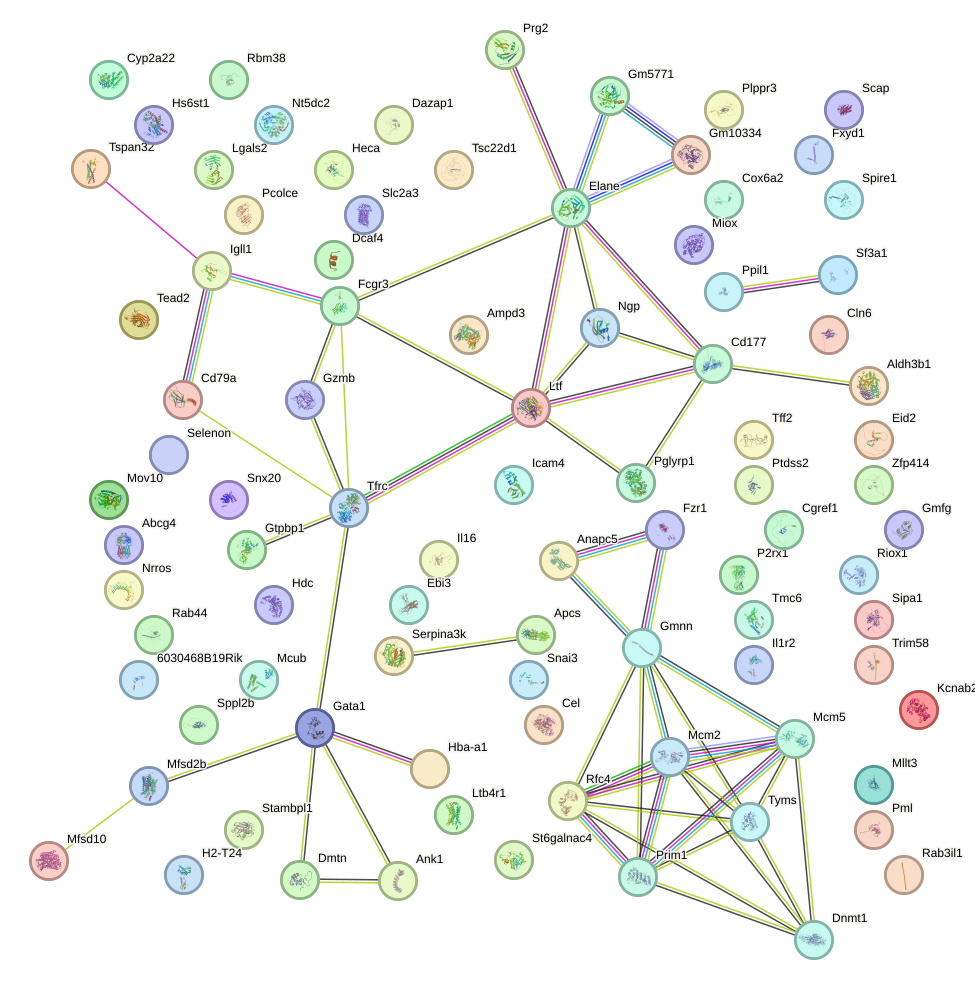
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarg
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_89218601 | 5.09 |
ENSMUST00000023282.9
|
Miox
|
myo-inositol oxygenase |
| chr7_-_142223662 | 4.19 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr7_+_18618605 | 3.73 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr9_+_20940669 | 3.66 |
ENSMUST00000001040.7
ENSMUST00000215077.2 |
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr2_+_172863688 | 3.61 |
ENSMUST00000029014.16
|
Rbm38
|
RNA binding motif protein 38 |
| chr10_+_79722081 | 3.61 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr9_+_110248815 | 3.59 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr6_+_41369290 | 3.56 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chrX_-_7834057 | 3.54 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr9_+_110848339 | 3.21 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr2_+_84810802 | 3.14 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr14_+_56003406 | 2.89 |
ENSMUST00000057569.4
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr7_-_24459736 | 2.81 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr7_+_140711181 | 2.80 |
ENSMUST00000026568.10
|
Ptdss2
|
phosphatidylserine synthase 2 |
| chr2_-_28453374 | 2.74 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr11_+_117688486 | 2.71 |
ENSMUST00000106331.2
|
6030468B19Rik
|
RIKEN cDNA 6030468B19 gene |
| chr17_+_29333116 | 2.68 |
ENSMUST00000233717.2
ENSMUST00000141239.2 |
Rab44
|
RAB44, member RAS oncogene family |
| chr7_+_44866635 | 2.44 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr14_-_70867588 | 2.42 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr7_-_126302315 | 2.30 |
ENSMUST00000173108.8
ENSMUST00000205515.2 |
Coro1a
|
coronin, actin binding protein 1A |
| chr10_+_128744689 | 2.25 |
ENSMUST00000105229.9
|
Cd63
|
CD63 antigen |
| chr11_+_117673107 | 2.25 |
ENSMUST00000050874.14
ENSMUST00000106334.9 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr7_-_127805518 | 2.21 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr12_-_4924341 | 2.20 |
ENSMUST00000137337.8
ENSMUST00000045921.14 |
Mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr5_-_30278552 | 2.17 |
ENSMUST00000198095.2
ENSMUST00000196872.2 ENSMUST00000026846.11 |
Tyms
|
thymidylate synthase |
| chr9_+_110173253 | 2.11 |
ENSMUST00000199709.3
|
Scap
|
SREBF chaperone |
| chr10_-_62178453 | 2.08 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr8_+_75836187 | 2.02 |
ENSMUST00000164309.3
ENSMUST00000212426.2 ENSMUST00000212811.2 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr7_+_142025575 | 1.99 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr11_+_78237492 | 1.97 |
ENSMUST00000100755.4
|
Unc119
|
unc-119 lipid binding chaperone |
| chr16_+_32427789 | 1.97 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr19_-_3979723 | 1.93 |
ENSMUST00000051803.8
|
Aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr11_+_32233511 | 1.89 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr1_-_170886924 | 1.86 |
ENSMUST00000164044.8
ENSMUST00000169017.8 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr1_-_172722589 | 1.86 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr17_+_47906985 | 1.86 |
ENSMUST00000182539.8
|
Ccnd3
|
cyclin D3 |
| chr18_-_67682312 | 1.85 |
ENSMUST00000224799.2
|
Spire1
|
spire type actin nucleation factor 1 |
| chr6_-_88875646 | 1.82 |
ENSMUST00000058011.8
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr9_-_20871081 | 1.81 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr7_+_24596806 | 1.79 |
ENSMUST00000003469.8
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr17_-_31363245 | 1.73 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr7_-_142209755 | 1.72 |
ENSMUST00000178921.2
|
Igf2
|
insulin-like growth factor 2 |
| chr17_+_56259617 | 1.70 |
ENSMUST00000003274.8
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr13_-_24945423 | 1.69 |
ENSMUST00000176890.8
ENSMUST00000175689.8 |
Gmnn
|
geminin |
| chr14_+_30856687 | 1.67 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr10_-_81214293 | 1.62 |
ENSMUST00000140901.8
|
Fzr1
|
fizzy and cell division cycle 20 related 1 |
| chr9_+_62754252 | 1.61 |
ENSMUST00000124984.2
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr11_-_117673008 | 1.56 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr16_-_22946441 | 1.53 |
ENSMUST00000133847.9
ENSMUST00000115338.8 ENSMUST00000023598.15 |
Rfc4
|
replication factor C (activator 1) 4 |
| chr7_+_28136861 | 1.51 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr2_+_157266175 | 1.50 |
ENSMUST00000029175.14
ENSMUST00000092576.11 |
Src
|
Rous sarcoma oncogene |
| chr16_+_32427738 | 1.50 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr9_-_44199832 | 1.49 |
ENSMUST00000161354.9
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chr17_-_28660688 | 1.45 |
ENSMUST00000233291.2
ENSMUST00000153744.2 |
Fkbp5
|
FK506 binding protein 5 |
| chr11_+_58531220 | 1.44 |
ENSMUST00000075084.5
|
Trim58
|
tripartite motif-containing 58 |
| chr4_-_134279433 | 1.43 |
ENSMUST00000060435.8
|
Selenon
|
selenoprotein N |
| chr10_-_62163444 | 1.42 |
ENSMUST00000139228.8
|
Hk1
|
hexokinase 1 |
| chr11_+_72889889 | 1.40 |
ENSMUST00000021141.14
|
P2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr12_-_114547622 | 1.39 |
ENSMUST00000193893.6
ENSMUST00000103498.3 |
Ighv1-9
|
immunoglobulin heavy variable V1-9 |
| chr13_-_22225527 | 1.37 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr5_-_31102829 | 1.36 |
ENSMUST00000031051.8
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr7_-_26638802 | 1.35 |
ENSMUST00000170227.3
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chr4_-_152533265 | 1.35 |
ENSMUST00000159840.8
ENSMUST00000105648.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr8_+_72050292 | 1.33 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr12_+_83997382 | 1.33 |
ENSMUST00000053744.9
|
Riox1
|
ribosomal oxygenase 1 |
| chr16_-_16681839 | 1.32 |
ENSMUST00000100136.4
|
Igll1
|
immunoglobulin lambda-like polypeptide 1 |
| chr19_+_9979033 | 1.32 |
ENSMUST00000121418.8
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr10_+_80691099 | 1.27 |
ENSMUST00000035597.10
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr17_-_36331416 | 1.25 |
ENSMUST00000174063.2
ENSMUST00000113760.10 |
H2-T24
|
histocompatibility 2, T region locus 24 |
| chr5_-_34794451 | 1.23 |
ENSMUST00000124668.2
ENSMUST00000001109.11 ENSMUST00000155577.8 ENSMUST00000114329.8 |
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr15_-_78739717 | 1.22 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr5_-_122959321 | 1.21 |
ENSMUST00000197074.5
ENSMUST00000199406.5 ENSMUST00000196640.5 ENSMUST00000197719.5 ENSMUST00000200645.5 |
Anapc5
|
anaphase-promoting complex subunit 5 |
| chr19_+_34194990 | 1.21 |
ENSMUST00000119603.2
|
Stambpl1
|
STAM binding protein like 1 |
| chr8_+_23629080 | 1.20 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr14_-_56499690 | 1.18 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr5_-_122959361 | 1.16 |
ENSMUST00000086216.9
|
Anapc5
|
anaphase-promoting complex subunit 5 |
| chr17_+_33848054 | 1.16 |
ENSMUST00000166627.8
ENSMUST00000073570.12 ENSMUST00000170225.3 |
Zfp414
|
zinc finger protein 414 |
| chr4_-_87724533 | 1.16 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr6_+_135339543 | 1.15 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr16_-_31984272 | 1.15 |
ENSMUST00000231836.2
ENSMUST00000115163.4 ENSMUST00000144345.2 ENSMUST00000143682.8 ENSMUST00000115165.10 ENSMUST00000099991.11 ENSMUST00000130410.8 |
Nrros
|
negative regulator of reactive oxygen species |
| chr3_-_129763638 | 1.15 |
ENSMUST00000146340.2
ENSMUST00000153506.8 |
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr17_-_29483075 | 1.13 |
ENSMUST00000024802.10
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr8_-_89362745 | 1.13 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr7_+_110368037 | 1.12 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr2_+_32477069 | 1.12 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr3_-_129763801 | 1.11 |
ENSMUST00000029624.15
|
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr4_-_152561896 | 1.09 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr1_+_40123858 | 1.09 |
ENSMUST00000027243.13
|
Il1r2
|
interleukin 1 receptor, type II |
| chr7_+_27967222 | 1.05 |
ENSMUST00000059596.8
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr7_+_142559375 | 1.04 |
ENSMUST00000075172.12
ENSMUST00000105923.8 |
Tspan32
|
tetraspanin 32 |
| chr3_-_104725853 | 1.04 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr7_-_83384711 | 1.04 |
ENSMUST00000001792.12
|
Il16
|
interleukin 16 |
| chr19_-_5713648 | 1.04 |
ENSMUST00000080824.13
ENSMUST00000237874.2 ENSMUST00000071857.13 ENSMUST00000236464.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr1_+_36107452 | 1.03 |
ENSMUST00000088174.4
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr15_+_79578141 | 1.01 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr14_+_76725876 | 1.01 |
ENSMUST00000101618.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr12_+_83572774 | 1.01 |
ENSMUST00000223291.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr12_-_113958518 | 1.00 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr2_-_126460575 | 1.00 |
ENSMUST00000028838.5
|
Hdc
|
histidine decarboxylase |
| chr12_-_114621406 | 1.00 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr9_-_58156982 | 0.99 |
ENSMUST00000135310.8
ENSMUST00000085673.11 ENSMUST00000114136.9 ENSMUST00000153820.8 ENSMUST00000124982.2 |
Pml
|
promyelocytic leukemia |
| chr10_+_127851031 | 0.99 |
ENSMUST00000178041.8
ENSMUST00000026461.8 |
Prim1
|
DNA primase, p49 subunit |
| chr11_+_4110346 | 0.99 |
ENSMUST00000002198.4
|
Sf3a1
|
splicing factor 3a, subunit 1 |
| chr8_-_123187406 | 0.98 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr5_-_137609634 | 0.98 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr10_+_80100812 | 0.98 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr6_-_122778598 | 0.97 |
ENSMUST00000165884.8
|
Slc2a3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr18_-_35795233 | 0.97 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr4_+_131600918 | 0.96 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr8_+_95712151 | 0.96 |
ENSMUST00000212799.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr2_+_103788321 | 0.96 |
ENSMUST00000156813.8
ENSMUST00000170926.8 |
Lmo2
|
LIM domain only 2 |
| chr10_-_68377672 | 0.96 |
ENSMUST00000020103.9
|
Cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr18_-_77855446 | 0.95 |
ENSMUST00000048192.9
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr8_+_71068791 | 0.95 |
ENSMUST00000210609.2
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr17_+_48539782 | 0.94 |
ENSMUST00000113251.10
ENSMUST00000048782.7 |
Trem1
|
triggering receptor expressed on myeloid cells 1 |
| chr5_-_137609691 | 0.94 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr19_-_46033353 | 0.94 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr5_+_137756407 | 0.94 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr7_+_30014235 | 0.93 |
ENSMUST00000054594.15
ENSMUST00000177078.8 ENSMUST00000176504.8 ENSMUST00000176304.8 |
Syne4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr5_-_34794546 | 0.92 |
ENSMUST00000114331.10
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr4_-_117740624 | 0.92 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr4_-_137493785 | 0.92 |
ENSMUST00000139951.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr17_-_48716756 | 0.91 |
ENSMUST00000160319.8
ENSMUST00000159535.2 ENSMUST00000078800.13 ENSMUST00000046719.14 ENSMUST00000162460.8 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr1_+_172328768 | 0.91 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr1_+_170472092 | 0.91 |
ENSMUST00000046792.9
|
Olfml2b
|
olfactomedin-like 2B |
| chr8_-_71725011 | 0.90 |
ENSMUST00000110071.3
|
Haus8
|
4HAUS augmin-like complex, subunit 8 |
| chr12_-_114140482 | 0.89 |
ENSMUST00000103475.2
ENSMUST00000195706.2 |
Ighv14-4
|
immunoglobulin heavy variable 14-4 |
| chr8_+_85696695 | 0.89 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr5_+_136067350 | 0.89 |
ENSMUST00000062606.8
|
Upk3b
|
uroplakin 3B |
| chr6_+_121187636 | 0.88 |
ENSMUST00000032233.9
|
Tuba8
|
tubulin, alpha 8 |
| chr15_+_6451721 | 0.88 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr19_-_5713701 | 0.88 |
ENSMUST00000164304.9
ENSMUST00000237544.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr6_+_71684846 | 0.88 |
ENSMUST00000212792.2
|
Reep1
|
receptor accessory protein 1 |
| chr7_+_126811831 | 0.87 |
ENSMUST00000127710.3
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr17_-_13213054 | 0.87 |
ENSMUST00000233867.2
|
Wtap
|
Wilms tumour 1-associating protein |
| chr16_+_33614378 | 0.87 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr19_-_4240984 | 0.85 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr17_+_29712008 | 0.84 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chr4_-_116228921 | 0.84 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr7_-_97827461 | 0.84 |
ENSMUST00000040971.14
|
Capn5
|
calpain 5 |
| chr9_-_58156935 | 0.84 |
ENSMUST00000124063.2
ENSMUST00000126690.8 |
Pml
|
promyelocytic leukemia |
| chr8_+_123829089 | 0.84 |
ENSMUST00000000756.6
|
Rpl13
|
ribosomal protein L13 |
| chr5_-_24935633 | 0.84 |
ENSMUST00000068693.12
|
Wdr86
|
WD repeat domain 86 |
| chr12_-_114752425 | 0.83 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr11_-_5753693 | 0.83 |
ENSMUST00000020768.4
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr14_+_66043281 | 0.83 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr7_+_43321426 | 0.83 |
ENSMUST00000038332.9
|
Ctu1
|
cytosolic thiouridylase subunit 1 |
| chr18_-_35795175 | 0.83 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr4_+_126915104 | 0.82 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr7_+_27151838 | 0.81 |
ENSMUST00000108357.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr8_+_23629046 | 0.81 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr16_+_23109213 | 0.81 |
ENSMUST00000115335.2
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr11_-_75060345 | 0.81 |
ENSMUST00000055619.5
|
Hic1
|
hypermethylated in cancer 1 |
| chr7_+_44117444 | 0.81 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr6_+_35154319 | 0.81 |
ENSMUST00000201374.4
ENSMUST00000043815.16 |
Nup205
|
nucleoporin 205 |
| chr11_+_117673198 | 0.81 |
ENSMUST00000117781.8
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr6_+_86348286 | 0.80 |
ENSMUST00000089558.7
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr14_-_54655079 | 0.80 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr17_-_27247581 | 0.78 |
ENSMUST00000143158.3
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr19_-_5713728 | 0.78 |
ENSMUST00000169854.2
|
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr9_+_119939414 | 0.76 |
ENSMUST00000035106.12
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr15_-_84441977 | 0.75 |
ENSMUST00000069476.5
|
Rtl6
|
retrotransposon Gag like 6 |
| chr3_-_83947416 | 0.75 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr6_+_135339929 | 0.75 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr6_-_83302890 | 0.74 |
ENSMUST00000204472.2
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr3_-_20209260 | 0.74 |
ENSMUST00000178328.8
|
Gyg
|
glycogenin |
| chr7_+_44117511 | 0.73 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr6_+_83003253 | 0.73 |
ENSMUST00000204891.2
|
M1ap
|
meiosis 1 associated protein |
| chr7_-_110581652 | 0.73 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr10_+_127871444 | 0.72 |
ENSMUST00000073868.9
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr16_+_31241085 | 0.72 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr17_-_33937565 | 0.72 |
ENSMUST00000174040.2
ENSMUST00000173015.8 ENSMUST00000066121.13 ENSMUST00000186022.7 ENSMUST00000173329.8 ENSMUST00000172767.9 |
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr11_+_76900091 | 0.71 |
ENSMUST00000129572.3
|
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr11_-_106890195 | 0.71 |
ENSMUST00000106768.2
ENSMUST00000144834.8 |
Kpna2
|
karyopherin (importin) alpha 2 |
| chr4_-_41314877 | 0.71 |
ENSMUST00000030145.9
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr10_+_128073900 | 0.71 |
ENSMUST00000105245.3
|
Timeless
|
timeless circadian clock 1 |
| chr15_-_103242697 | 0.70 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr19_-_4241034 | 0.70 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr14_+_71127540 | 0.70 |
ENSMUST00000022699.10
|
Gfra2
|
glial cell line derived neurotrophic factor family receptor alpha 2 |
| chr10_+_79650496 | 0.70 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr7_+_3340013 | 0.70 |
ENSMUST00000204541.2
|
Myadm
|
myeloid-associated differentiation marker |
| chr19_+_5530073 | 0.69 |
ENSMUST00000164388.2
|
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr2_-_155424576 | 0.69 |
ENSMUST00000126322.8
|
Gss
|
glutathione synthetase |
| chr4_+_109272828 | 0.69 |
ENSMUST00000106618.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr3_-_20209220 | 0.69 |
ENSMUST00000184552.2
|
Gyg
|
glycogenin |
| chr6_-_122587005 | 0.69 |
ENSMUST00000032211.5
|
Gdf3
|
growth differentiation factor 3 |
| chr7_+_44117404 | 0.68 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr9_-_107960528 | 0.68 |
ENSMUST00000159372.3
ENSMUST00000160249.8 |
Rnf123
|
ring finger protein 123 |
| chr7_+_44117475 | 0.68 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr13_-_56326511 | 0.68 |
ENSMUST00000169652.3
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr9_+_20927271 | 0.68 |
ENSMUST00000086399.6
|
Icam1
|
intercellular adhesion molecule 1 |
| chr1_+_63215976 | 0.68 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_+_24852409 | 0.68 |
ENSMUST00000028351.9
|
Dph7
|
diphthamine biosynethesis 7 |
| chr11_-_106890307 | 0.67 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr5_-_113163339 | 0.67 |
ENSMUST00000197776.2
ENSMUST00000065167.9 |
Grk3
|
G protein-coupled receptor kinase 3 |
| chr7_-_45044606 | 0.67 |
ENSMUST00000209858.2
|
Snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr10_+_122514669 | 0.66 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr3_-_96201248 | 0.66 |
ENSMUST00000029748.8
|
Fcgr1
|
Fc receptor, IgG, high affinity I |
| chr12_-_115122455 | 0.65 |
ENSMUST00000103523.2
|
Ighv1-53
|
immunoglobulin heavy variable 1-53 |
| chr11_-_82799186 | 0.65 |
ENSMUST00000103213.10
|
Nle1
|
notchless homolog 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 1.2 | 3.6 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) neutrophil mediated killing of fungus(GO:0070947) |
| 1.2 | 3.5 | GO:0030221 | basophil differentiation(GO:0030221) |
| 1.0 | 3.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.9 | 3.6 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.8 | 2.5 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.8 | 3.2 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.7 | 2.6 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.6 | 1.9 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.5 | 2.7 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.5 | 1.5 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.5 | 1.5 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.5 | 2.0 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.5 | 1.8 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.5 | 1.8 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 | 1.3 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.4 | 3.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 2.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.4 | 2.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 5.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.4 | 2.2 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.4 | 2.2 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.4 | 1.4 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.4 | 1.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.3 | 2.7 | GO:0032796 | uropod organization(GO:0032796) |
| 0.3 | 1.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.3 | 1.0 | GO:0097037 | heme export(GO:0097037) |
| 0.3 | 0.9 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 5.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 0.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 0.3 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 | 1.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.3 | 1.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 1.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.5 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.2 | 1.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 0.7 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 1.4 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.2 | 1.4 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 | 1.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 0.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 3.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 1.0 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.2 | 2.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 0.8 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.2 | 1.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.8 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 0.5 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 0.7 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.2 | 0.5 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 1.5 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 0.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 1.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.8 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 1.3 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 0.5 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 | 1.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 1.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 1.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 1.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.9 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.7 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.1 | 1.6 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 1.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.6 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.7 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 1.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.8 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.4 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.7 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 3.0 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.1 | 3.3 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.1 | 1.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.5 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.1 | 0.7 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.8 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 1.6 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 1.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 1.1 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.4 | GO:0035962 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) interleukin-4-mediated signaling pathway(GO:0035771) response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 0.7 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 16.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.9 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.1 | 1.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.7 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.3 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.1 | 1.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 1.9 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 3.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.7 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.4 | GO:1903943 | estrous cycle(GO:0044849) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.5 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.8 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.9 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 0.3 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) spermidine catabolic process(GO:0046203) |
| 0.1 | 0.4 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 2.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.8 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.3 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.5 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 1.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 1.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.2 | GO:0042628 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) seminal vesicle development(GO:0061107) |
| 0.1 | 0.4 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.3 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.3 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.9 | GO:0050755 | chemokine metabolic process(GO:0050755) |
| 0.1 | 3.4 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 0.3 | GO:0060733 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.1 | 0.6 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 2.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.9 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.6 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 2.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 1.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 2.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.6 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 0.4 | GO:0061198 | fungiform papilla formation(GO:0061198) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 1.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 0.8 | GO:1903140 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.1 | 0.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.3 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.1 | 0.4 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 0.5 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.1 | 0.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.5 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 1.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 2.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 1.1 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 1.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.5 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.6 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.6 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.4 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.0 | 0.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.5 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.8 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) |
| 0.0 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 1.0 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 2.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.5 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 1.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 1.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.7 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.2 | GO:1905216 | regulation of mRNA binding(GO:1902415) positive regulation of mRNA binding(GO:1902416) regulation of RNA binding(GO:1905214) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.3 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.4 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 1.0 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.4 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.9 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.5 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 2.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:1905153 | regulation of phagocytosis, engulfment(GO:0060099) regulation of membrane invagination(GO:1905153) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.9 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.5 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.7 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.6 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.3 | GO:0071548 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 1.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.4 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.2 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.6 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.8 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.7 | 2.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.6 | 2.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.5 | 3.2 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.5 | 1.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.5 | 1.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.4 | 1.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.4 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.3 | 2.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 1.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 2.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 1.6 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.3 | 1.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 1.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 3.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 3.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.3 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 1.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 1.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 4.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.2 | 1.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 0.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 0.5 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 0.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 0.2 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.1 | 16.2 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 4.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.9 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 0.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 2.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 1.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 2.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 3.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 2.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 2.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 2.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.2 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 7.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 3.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 2.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.0 | GO:0030684 | preribosome(GO:0030684) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 1.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.7 | 3.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.7 | 2.6 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.6 | 2.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.5 | 3.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.5 | 1.9 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 3.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 2.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 1.9 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.4 | 4.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 1.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.3 | 0.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.3 | 0.8 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 0.8 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.3 | 1.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 1.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.3 | 1.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 2.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 3.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 1.0 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.2 | 0.7 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.2 | 0.5 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.2 | 2.8 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 1.9 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 3.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 0.6 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 1.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 3.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 0.6 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.2 | 1.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.3 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 1.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 1.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 1.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 1.9 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 1.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.4 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.7 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 1.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.7 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 1.9 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 1.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 1.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 16.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 1.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.3 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 0.6 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 4.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.6 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.7 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 1.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 2.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 1.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 2.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.2 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 2.6 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.8 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 2.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 4.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 2.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 1.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.6 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 1.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 5.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 1.0 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 6.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 4.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 2.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.6 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 2.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 5.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.9 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.2 | 1.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.8 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.1 | 3.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 4.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 0.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 4.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 2.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 2.0 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 1.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 2.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.4 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |