Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
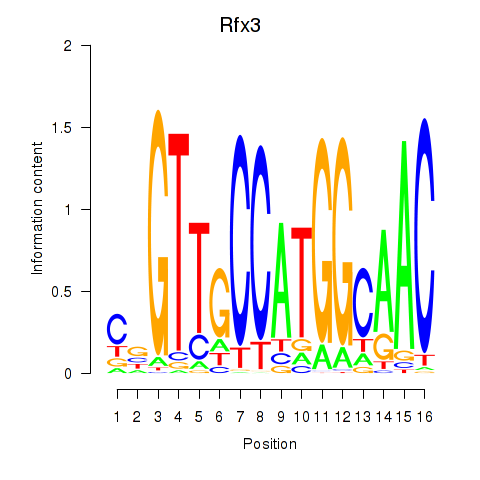
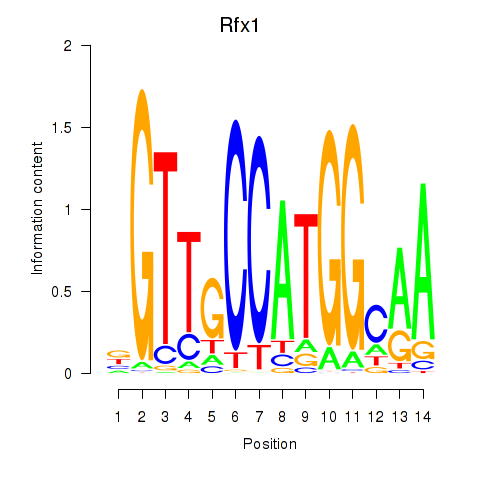
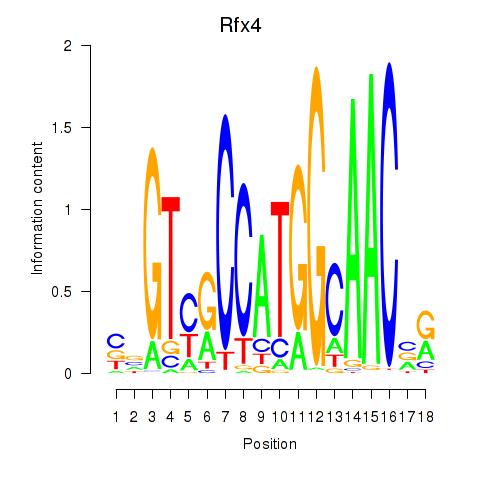
Results for Rfx3_Rfx1_Rfx4
Z-value: 1.81
Transcription factors associated with Rfx3_Rfx1_Rfx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx3
|
ENSMUSG00000040929.18 | Rfx3 |
|
Rfx1
|
ENSMUSG00000031706.8 | Rfx1 |
|
Rfx4
|
ENSMUSG00000020037.16 | Rfx4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx3 | mm39_v1_chr19_-_27988393_27988453 | -0.67 | 7.2e-06 | Click! |
| Rfx1 | mm39_v1_chr8_+_84793453_84793511 | -0.57 | 2.7e-04 | Click! |
| Rfx4 | mm39_v1_chr10_+_84674008_84674016 | 0.49 | 2.7e-03 | Click! |
Activity profile of Rfx3_Rfx1_Rfx4 motif
Sorted Z-values of Rfx3_Rfx1_Rfx4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx3_Rfx1_Rfx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_5900019 | 10.68 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr11_+_101358990 | 6.07 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr5_+_30972067 | 4.65 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr5_+_30971915 | 4.61 |
ENSMUST00000031058.15
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr10_-_39901249 | 3.98 |
ENSMUST00000163705.3
|
Mfsd4b1
|
major facilitator superfamily domain containing 4B1 |
| chr7_+_44114815 | 3.52 |
ENSMUST00000035929.11
ENSMUST00000146128.8 |
Aspdh
|
aspartate dehydrogenase domain containing |
| chr12_+_113104085 | 3.43 |
ENSMUST00000200380.5
|
Crip2
|
cysteine rich protein 2 |
| chr1_-_192946359 | 3.38 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr7_+_44114857 | 3.26 |
ENSMUST00000135624.2
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr7_-_30754193 | 3.23 |
ENSMUST00000205778.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_30754223 | 3.15 |
ENSMUST00000206012.2
ENSMUST00000108110.5 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_30754240 | 3.11 |
ENSMUST00000206860.2
ENSMUST00000071697.11 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr12_+_113103817 | 3.10 |
ENSMUST00000084882.9
|
Crip2
|
cysteine rich protein 2 |
| chr9_+_118931532 | 3.09 |
ENSMUST00000165231.8
ENSMUST00000140326.8 |
Dlec1
|
deleted in lung and esophageal cancer 1 |
| chr4_-_43669141 | 2.96 |
ENSMUST00000056474.7
|
Fam221b
|
family with sequence similarity 221, member B |
| chr9_-_106563002 | 2.93 |
ENSMUST00000163441.8
|
Tex264
|
testis expressed gene 264 |
| chr11_-_105835238 | 2.92 |
ENSMUST00000019734.11
ENSMUST00000184269.3 ENSMUST00000150563.3 |
Cyb561
|
cytochrome b-561 |
| chr9_-_60595401 | 2.91 |
ENSMUST00000114034.9
ENSMUST00000065603.12 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr9_-_106562852 | 2.86 |
ENSMUST00000169068.8
|
Tex264
|
testis expressed gene 264 |
| chr7_-_30754792 | 2.84 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr15_-_85918378 | 2.70 |
ENSMUST00000016172.10
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr17_-_34822649 | 2.69 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr8_-_3517617 | 2.67 |
ENSMUST00000111081.10
ENSMUST00000004686.13 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr9_-_106563091 | 2.67 |
ENSMUST00000046735.11
|
Tex264
|
testis expressed gene 264 |
| chr17_-_47063095 | 2.63 |
ENSMUST00000121671.2
ENSMUST00000059844.13 |
Cnpy3
|
canopy FGF signaling regulator 3 |
| chr3_-_89121686 | 2.62 |
ENSMUST00000073572.12
|
Mtx1
|
metaxin 1 |
| chr9_-_72892617 | 2.58 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr2_-_90735171 | 2.56 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr16_-_3726503 | 2.53 |
ENSMUST00000115859.8
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr4_+_124696394 | 2.43 |
ENSMUST00000175875.2
|
Mtf1
|
metal response element binding transcription factor 1 |
| chr7_-_126522014 | 2.42 |
ENSMUST00000134134.3
ENSMUST00000119781.8 ENSMUST00000121612.4 |
Tmem219
|
transmembrane protein 219 |
| chr9_+_108924457 | 2.40 |
ENSMUST00000072093.13
|
Plxnb1
|
plexin B1 |
| chr3_-_89121637 | 2.39 |
ENSMUST00000239452.2
ENSMUST00000118964.10 |
Mtx1
|
metaxin 1 |
| chr2_-_5719302 | 2.38 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr17_-_56916771 | 2.28 |
ENSMUST00000052832.6
|
Micos13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr11_+_108316018 | 2.24 |
ENSMUST00000150863.9
ENSMUST00000061287.12 ENSMUST00000149683.9 |
Cep112
|
centrosomal protein 112 |
| chr12_+_87193922 | 2.14 |
ENSMUST00000222885.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr9_+_59496571 | 2.10 |
ENSMUST00000121266.8
ENSMUST00000118164.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr1_-_133849131 | 2.08 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr3_-_54962899 | 2.07 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr15_+_76580925 | 2.07 |
ENSMUST00000023203.6
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr9_+_107926502 | 2.07 |
ENSMUST00000047947.9
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr11_-_4110286 | 2.06 |
ENSMUST00000093381.11
ENSMUST00000101626.9 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr3_-_54962922 | 2.05 |
ENSMUST00000197238.5
|
Ccna1
|
cyclin A1 |
| chr17_+_26048010 | 2.03 |
ENSMUST00000026832.14
|
Jmjd8
|
jumonji domain containing 8 |
| chr10_-_41366321 | 2.03 |
ENSMUST00000019965.13
|
Smpd2
|
sphingomyelin phosphodiesterase 2, neutral |
| chr4_+_152423344 | 2.03 |
ENSMUST00000005175.5
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr17_-_24915121 | 2.01 |
ENSMUST00000046839.10
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr6_-_122317156 | 1.98 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr18_-_39051695 | 1.90 |
ENSMUST00000040647.11
|
Fgf1
|
fibroblast growth factor 1 |
| chr7_+_44078366 | 1.90 |
ENSMUST00000127790.8
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr4_-_155753628 | 1.90 |
ENSMUST00000103176.10
|
Mib2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr15_+_76555815 | 1.87 |
ENSMUST00000037551.15
|
Ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr10_+_34173426 | 1.87 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr8_+_123939596 | 1.84 |
ENSMUST00000212892.2
ENSMUST00000212161.2 ENSMUST00000060133.8 ENSMUST00000212346.2 ENSMUST00000212637.2 |
Spata33
|
spermatogenesis associated 33 |
| chr12_-_86773160 | 1.83 |
ENSMUST00000021682.9
|
Angel1
|
angel homolog 1 |
| chr17_+_46565116 | 1.83 |
ENSMUST00000095262.6
|
Lrrc73
|
leucine rich repeat containing 73 |
| chr7_+_18725170 | 1.83 |
ENSMUST00000059331.9
ENSMUST00000131087.2 |
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr8_+_123939566 | 1.82 |
ENSMUST00000212760.2
ENSMUST00000212523.2 |
Spata33
|
spermatogenesis associated 33 |
| chr4_+_43669266 | 1.81 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr9_+_110306020 | 1.78 |
ENSMUST00000198858.5
|
Kif9
|
kinesin family member 9 |
| chr9_-_121668527 | 1.76 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr10_-_81308693 | 1.76 |
ENSMUST00000147524.3
ENSMUST00000119060.8 |
Celf5
|
CUGBP, Elav-like family member 5 |
| chr7_-_79392763 | 1.75 |
ENSMUST00000032761.8
|
Pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr8_-_65186565 | 1.74 |
ENSMUST00000141021.2
|
Msmo1
|
methylsterol monoxygenase 1 |
| chr15_+_76555838 | 1.74 |
ENSMUST00000135388.3
|
Ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr2_+_121120070 | 1.74 |
ENSMUST00000094639.10
|
Map1a
|
microtubule-associated protein 1 A |
| chr11_+_102036356 | 1.73 |
ENSMUST00000055409.6
|
Nags
|
N-acetylglutamate synthase |
| chr2_+_145776720 | 1.72 |
ENSMUST00000152515.8
ENSMUST00000138774.8 ENSMUST00000130168.8 ENSMUST00000133433.8 ENSMUST00000118002.2 |
Cfap61
|
cilia and flagella associated protein 61 |
| chr9_+_108447077 | 1.70 |
ENSMUST00000019183.14
|
Dalrd3
|
DALR anticodon binding domain containing 3 |
| chr17_+_35055962 | 1.69 |
ENSMUST00000173874.8
ENSMUST00000180043.8 ENSMUST00000046244.15 |
Dxo
|
decapping exoribonuclease |
| chr9_+_74860133 | 1.68 |
ENSMUST00000215370.2
|
Fam214a
|
family with sequence similarity 214, member A |
| chr9_-_63509699 | 1.68 |
ENSMUST00000171243.2
ENSMUST00000163982.8 ENSMUST00000163624.8 |
Iqch
|
IQ motif containing H |
| chr17_+_46558995 | 1.66 |
ENSMUST00000095263.10
ENSMUST00000123311.8 |
Yipf3
|
Yip1 domain family, member 3 |
| chr9_-_70048766 | 1.65 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr3_+_103875904 | 1.64 |
ENSMUST00000156262.8
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr13_-_58506890 | 1.64 |
ENSMUST00000225388.2
|
Kif27
|
kinesin family member 27 |
| chrX_-_50294652 | 1.63 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr4_+_130001349 | 1.63 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr7_-_133378468 | 1.62 |
ENSMUST00000033290.12
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr4_-_119272690 | 1.61 |
ENSMUST00000238287.2
ENSMUST00000238759.2 ENSMUST00000063642.10 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr4_+_41465134 | 1.61 |
ENSMUST00000030154.7
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr17_+_80514889 | 1.61 |
ENSMUST00000134652.2
|
Ttc39d
|
tetratricopeptide repeat domain 39D |
| chr7_-_133378410 | 1.61 |
ENSMUST00000130182.2
ENSMUST00000106139.8 |
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr17_-_24915037 | 1.60 |
ENSMUST00000234235.2
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr2_+_145776697 | 1.58 |
ENSMUST00000116398.8
ENSMUST00000126415.8 |
Cfap61
|
cilia and flagella associated protein 61 |
| chr17_+_85928459 | 1.56 |
ENSMUST00000162695.3
|
Six3
|
sine oculis-related homeobox 3 |
| chr7_+_27878894 | 1.54 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr3_+_94391676 | 1.54 |
ENSMUST00000198384.3
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr5_-_142536720 | 1.53 |
ENSMUST00000129212.2
ENSMUST00000110785.2 ENSMUST00000063635.15 |
Radil
|
Ras association and DIL domains |
| chr7_+_27879650 | 1.53 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr11_-_105834953 | 1.53 |
ENSMUST00000184086.8
|
Cyb561
|
cytochrome b-561 |
| chr9_+_107926441 | 1.52 |
ENSMUST00000112295.9
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr7_+_3439144 | 1.49 |
ENSMUST00000182222.8
|
Cacng8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr9_+_47441471 | 1.49 |
ENSMUST00000114548.8
ENSMUST00000152459.8 ENSMUST00000143026.9 ENSMUST00000085909.9 ENSMUST00000114547.8 ENSMUST00000239368.2 ENSMUST00000214542.2 ENSMUST00000034581.4 |
Cadm1
|
cell adhesion molecule 1 |
| chr6_-_72496986 | 1.47 |
ENSMUST00000160123.9
ENSMUST00000234038.2 ENSMUST00000235004.2 |
Sh2d6
|
SH2 domain containing 6 |
| chr14_-_25927674 | 1.46 |
ENSMUST00000100811.6
|
Tmem254a
|
transmembrane protein 254a |
| chr9_+_110306052 | 1.46 |
ENSMUST00000197248.5
ENSMUST00000061155.12 ENSMUST00000198043.5 ENSMUST00000084952.8 |
Kif9
|
kinesin family member 9 |
| chr19_+_5100815 | 1.46 |
ENSMUST00000224178.2
ENSMUST00000225799.3 ENSMUST00000025818.8 |
Rin1
|
Ras and Rab interactor 1 |
| chr9_-_63509747 | 1.45 |
ENSMUST00000080527.12
ENSMUST00000042322.11 |
Iqch
|
IQ motif containing H |
| chr2_+_153334710 | 1.45 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr3_+_94391644 | 1.44 |
ENSMUST00000197677.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr4_-_119272667 | 1.44 |
ENSMUST00000238609.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr12_-_84455764 | 1.43 |
ENSMUST00000120942.8
ENSMUST00000110272.9 |
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr19_-_45548942 | 1.42 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chr11_-_59730654 | 1.42 |
ENSMUST00000019517.10
|
Cops3
|
COP9 signalosome subunit 3 |
| chr19_+_45549009 | 1.42 |
ENSMUST00000047057.9
|
Gm17018
|
predicted gene 17018 |
| chr19_+_5100475 | 1.41 |
ENSMUST00000225427.2
|
Rin1
|
Ras and Rab interactor 1 |
| chr9_+_74860335 | 1.41 |
ENSMUST00000170846.8
|
Fam214a
|
family with sequence similarity 214, member A |
| chr11_-_101316156 | 1.40 |
ENSMUST00000103102.10
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr5_-_87240405 | 1.40 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr5_+_30869193 | 1.40 |
ENSMUST00000088081.11
ENSMUST00000101442.4 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr2_-_26030453 | 1.38 |
ENSMUST00000133808.2
|
Tmem250-ps
|
transmembrane protein 250, pseudogene |
| chr11_-_105347500 | 1.38 |
ENSMUST00000049995.10
ENSMUST00000100332.4 |
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr9_+_50528813 | 1.38 |
ENSMUST00000141366.8
|
Pih1d2
|
PIH1 domain containing 2 |
| chr9_-_21913833 | 1.37 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr7_+_29931309 | 1.37 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr5_+_138170259 | 1.36 |
ENSMUST00000019662.11
ENSMUST00000151318.8 |
Ap4m1
|
adaptor-related protein complex AP-4, mu 1 |
| chr10_+_80134917 | 1.36 |
ENSMUST00000154212.8
|
Apc2
|
APC regulator of WNT signaling pathway 2 |
| chr15_+_100659729 | 1.35 |
ENSMUST00000161564.2
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr19_+_6413703 | 1.34 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr7_-_12788441 | 1.34 |
ENSMUST00000182087.2
|
Mzf1
|
myeloid zinc finger 1 |
| chr10_-_39775182 | 1.34 |
ENSMUST00000178045.9
ENSMUST00000178563.3 |
Mfsd4b4
|
major facilitator superfamily domain containing 4B4 |
| chr9_-_21913896 | 1.34 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr7_+_29931735 | 1.34 |
ENSMUST00000108193.2
ENSMUST00000108192.2 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr6_+_30509826 | 1.34 |
ENSMUST00000031797.11
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr12_-_104010690 | 1.33 |
ENSMUST00000043915.4
|
Serpina12
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr13_+_43769426 | 1.33 |
ENSMUST00000161817.2
|
Rnf182
|
ring finger protein 182 |
| chr15_-_89064936 | 1.33 |
ENSMUST00000109331.9
|
Plxnb2
|
plexin B2 |
| chr16_-_90866032 | 1.32 |
ENSMUST00000035689.8
ENSMUST00000114076.2 |
4932438H23Rik
|
RIKEN cDNA 4932438H23 gene |
| chr5_+_30868908 | 1.32 |
ENSMUST00000114729.8
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr6_-_83102010 | 1.32 |
ENSMUST00000032109.5
|
Ino80b
|
INO80 complex subunit B |
| chr9_+_43978290 | 1.31 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr7_+_6733561 | 1.31 |
ENSMUST00000200535.6
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr16_-_3726452 | 1.31 |
ENSMUST00000123235.3
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr2_-_73605684 | 1.31 |
ENSMUST00000112024.10
ENSMUST00000180045.8 |
Chn1
|
chimerin 1 |
| chr7_+_45434755 | 1.30 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr19_-_4889284 | 1.30 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr17_+_57318469 | 1.29 |
ENSMUST00000210548.2
|
Gm17949
|
predicted gene, 17949 |
| chr3_-_89121147 | 1.29 |
ENSMUST00000173477.8
ENSMUST00000119222.9 |
Mtx1
|
metaxin 1 |
| chr10_-_80895949 | 1.29 |
ENSMUST00000219401.2
ENSMUST00000220317.2 ENSMUST00000005067.6 ENSMUST00000218208.2 |
Sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr19_-_4889314 | 1.28 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr4_+_152423075 | 1.28 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr17_-_36290129 | 1.28 |
ENSMUST00000165613.9
ENSMUST00000173872.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr6_+_34757346 | 1.27 |
ENSMUST00000115016.8
ENSMUST00000115017.8 |
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr7_-_118304930 | 1.27 |
ENSMUST00000207323.2
ENSMUST00000038791.15 |
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr17_-_26014613 | 1.27 |
ENSMUST00000235889.2
|
Gm50367
|
predicted gene, 50367 |
| chr11_+_101443014 | 1.27 |
ENSMUST00000147239.8
|
Nbr1
|
NBR1, autophagy cargo receptor |
| chr13_+_43769186 | 1.26 |
ENSMUST00000059986.3
|
Rnf182
|
ring finger protein 182 |
| chr5_-_149559667 | 1.25 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr15_-_76010736 | 1.25 |
ENSMUST00000054022.12
ENSMUST00000089654.4 |
BC024139
|
cDNA sequence BC024139 |
| chr7_+_6175532 | 1.25 |
ENSMUST00000207329.2
|
Zfp444
|
zinc finger protein 444 |
| chr5_+_120569764 | 1.25 |
ENSMUST00000031591.10
|
Lhx5
|
LIM homeobox protein 5 |
| chr2_-_35939377 | 1.25 |
ENSMUST00000070112.6
|
Ndufa8
|
NADH:ubiquinone oxidoreductase subunit A8 |
| chr10_+_87695117 | 1.24 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr2_-_73605387 | 1.23 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr12_-_103739847 | 1.23 |
ENSMUST00000078869.6
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr4_+_138161958 | 1.23 |
ENSMUST00000044058.11
ENSMUST00000105813.8 ENSMUST00000105815.2 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr12_-_83609217 | 1.22 |
ENSMUST00000222448.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr18_-_78249612 | 1.22 |
ENSMUST00000163367.3
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr3_-_89152320 | 1.22 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr4_-_102971752 | 1.22 |
ENSMUST00000106868.4
|
Dnai4
|
dynein axonemal intermediate chain 4 |
| chr5_-_149559792 | 1.20 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr4_-_102971434 | 1.20 |
ENSMUST00000036557.15
ENSMUST00000036451.15 |
Dnai4
|
dynein axonemal intermediate chain 4 |
| chr7_-_79882228 | 1.20 |
ENSMUST00000123279.8
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr19_+_6111204 | 1.19 |
ENSMUST00000162726.5
|
Znhit2
|
zinc finger, HIT domain containing 2 |
| chr17_-_45903494 | 1.17 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr4_-_141742367 | 1.17 |
ENSMUST00000105780.8
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr17_-_85397627 | 1.17 |
ENSMUST00000072406.5
ENSMUST00000171795.9 ENSMUST00000234676.2 |
Prepl
|
prolyl endopeptidase-like |
| chr10_+_106306122 | 1.17 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr5_+_123390149 | 1.16 |
ENSMUST00000121964.8
|
Wdr66
|
WD repeat domain 66 |
| chrX_+_7786061 | 1.15 |
ENSMUST00000041096.4
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr9_-_121106209 | 1.15 |
ENSMUST00000051479.13
ENSMUST00000171923.8 |
Ulk4
|
unc-51-like kinase 4 |
| chr6_+_55180366 | 1.15 |
ENSMUST00000204842.3
ENSMUST00000053094.8 |
Mindy4
|
MINDY lysine 48 deubiquitinase 4 |
| chr13_+_25029088 | 1.15 |
ENSMUST00000006893.9
|
D130043K22Rik
|
RIKEN cDNA D130043K22 gene |
| chr5_+_29400981 | 1.14 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr9_+_72892693 | 1.14 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr14_-_54491365 | 1.12 |
ENSMUST00000128231.2
|
Dad1
|
defender against cell death 1 |
| chr11_-_116226175 | 1.12 |
ENSMUST00000036215.8
|
Foxj1
|
forkhead box J1 |
| chr7_-_29426425 | 1.11 |
ENSMUST00000061193.4
|
Catsperg2
|
cation channel sperm associated auxiliary subunit gamma 2 |
| chr7_+_108549545 | 1.11 |
ENSMUST00000207583.2
|
Tub
|
tubby bipartite transcription factor |
| chr6_+_55994473 | 1.10 |
ENSMUST00000052827.6
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr5_+_66417488 | 1.09 |
ENSMUST00000031109.7
|
Nsun7
|
NOL1/NOP2/Sun domain family, member 7 |
| chr7_+_98917542 | 1.09 |
ENSMUST00000107100.3
ENSMUST00000208605.2 |
Map6
|
microtubule-associated protein 6 |
| chr17_-_36290571 | 1.09 |
ENSMUST00000173724.2
ENSMUST00000172900.8 ENSMUST00000174849.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr8_+_46080840 | 1.09 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr16_-_95993420 | 1.08 |
ENSMUST00000113804.8
ENSMUST00000054855.14 |
Lca5l
|
Leber congenital amaurosis 5-like |
| chr3_+_82265351 | 1.08 |
ENSMUST00000193559.6
ENSMUST00000192595.6 ENSMUST00000091014.10 |
Map9
|
microtubule-associated protein 9 |
| chr10_+_42736539 | 1.07 |
ENSMUST00000157071.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr9_-_24414423 | 1.07 |
ENSMUST00000142064.8
ENSMUST00000170356.2 |
Dpy19l1
|
dpy-19-like 1 (C. elegans) |
| chr2_-_164699462 | 1.07 |
ENSMUST00000109316.8
ENSMUST00000156255.8 ENSMUST00000128110.2 ENSMUST00000109317.10 ENSMUST00000059954.14 |
Pltp
|
phospholipid transfer protein |
| chr4_-_119272640 | 1.07 |
ENSMUST00000238293.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr7_+_6733684 | 1.06 |
ENSMUST00000197117.5
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr7_-_140462187 | 1.06 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr9_-_64248426 | 1.06 |
ENSMUST00000120760.8
ENSMUST00000168844.9 |
Dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr1_-_60082246 | 1.06 |
ENSMUST00000027172.13
ENSMUST00000191251.7 |
Ica1l
|
islet cell autoantigen 1-like |
| chr2_+_35939463 | 1.06 |
ENSMUST00000028256.5
|
Morn5
|
MORN repeat containing 5 |
| chr17_-_56424577 | 1.05 |
ENSMUST00000019808.12
|
Plin5
|
perilipin 5 |
| chr9_-_51013361 | 1.05 |
ENSMUST00000170947.3
|
Hoatz
|
HOATZ cilia and flagella associated protein |
| chr2_+_164302863 | 1.04 |
ENSMUST00000072452.11
|
Sys1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.3 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 2.1 | 10.7 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 1.1 | 6.8 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 1.1 | 3.4 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.1 | 12.3 | GO:1903797 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 1.0 | 1.0 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.9 | 2.7 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.9 | 4.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.8 | 2.3 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.7 | 2.7 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.6 | 2.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 | 1.6 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.5 | 1.6 | GO:1904733 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.5 | 1.6 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.5 | 1.9 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.5 | 1.9 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.5 | 1.8 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 1.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.4 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.4 | 3.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.4 | 2.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.4 | 2.4 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.4 | 2.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.4 | 1.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 1.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.4 | 1.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.4 | 6.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 1.1 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.4 | 1.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 3.5 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.3 | 1.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.3 | 1.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 1.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.3 | 3.6 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.3 | 2.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 1.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.3 | 2.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 2.1 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.3 | 2.8 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.3 | 1.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 1.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.3 | 1.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 0.8 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.3 | 1.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 0.5 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.2 | 0.7 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 2.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 2.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 1.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 0.7 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 2.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.7 | GO:0071609 | chemokine (C-C motif) ligand 5 production(GO:0071609) |
| 0.2 | 1.2 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.2 | 2.0 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.2 | 1.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 2.5 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 0.8 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.2 | 1.0 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 0.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 3.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.2 | 2.6 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.2 | 2.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 2.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.5 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 1.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.5 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.9 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.4 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 2.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.8 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 1.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.9 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.4 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 1.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.1 | 2.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 2.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 1.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.5 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 1.4 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 3.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 3.0 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 4.9 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 3.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.4 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.5 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.7 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.7 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 3.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.1 | 1.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 2.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.0 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 3.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 1.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 5.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.3 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.1 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 1.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 2.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.3 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 | 0.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.3 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 2.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.4 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.1 | 0.8 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.0 | 1.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 2.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 1.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 3.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.8 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 2.1 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 1.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0039526 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 1.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.2 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.3 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 2.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.8 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.4 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.9 | GO:2000191 | regulation of fatty acid transport(GO:2000191) |
| 0.0 | 0.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 2.5 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 2.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 2.3 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 3.7 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.7 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 2.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.8 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 1.0 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.6 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.8 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 2.7 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.4 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.4 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 1.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 1.9 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 1.5 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.6 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.0 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.6 | 10.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 2.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.5 | 1.8 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.3 | 0.9 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 1.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 0.9 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 0.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 0.9 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 2.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 2.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 1.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 7.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 2.0 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 2.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 2.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 4.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.2 | 2.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 0.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 2.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 10.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 3.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 2.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 2.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 4.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 2.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.3 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.1 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 2.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.2 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 2.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 1.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 1.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 2.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 2.0 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 2.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.5 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 1.1 | 3.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.1 | 10.7 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.8 | 3.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.7 | 3.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.7 | 2.1 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.5 | 2.7 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.4 | 2.1 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.4 | 2.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 1.6 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.3 | 1.7 | GO:0034618 | arginine binding(GO:0034618) |
| 0.3 | 1.7 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.3 | 1.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.3 | 3.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 0.9 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.3 | 3.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 1.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.3 | 1.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.3 | 1.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 3.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 1.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 6.8 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.2 | 1.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 0.7 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.2 | 1.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 2.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 0.5 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 1.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 1.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.9 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 5.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 2.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 2.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 1.3 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.4 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 1.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.5 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.9 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 7.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 2.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 2.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 2.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 3.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 0.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 2.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.2 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 1.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.2 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 2.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 2.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 2.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.8 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.3 | GO:0071209 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 2.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0070061 | fructose-bisphosphate aldolase activity(GO:0004332) fructose binding(GO:0070061) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.0 | 2.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 1.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 1.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 2.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 5.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 2.5 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 5.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 1.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 2.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.5 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.1 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 10.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 3.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 5.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 11.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 2.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 3.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 2.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 4.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |