Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
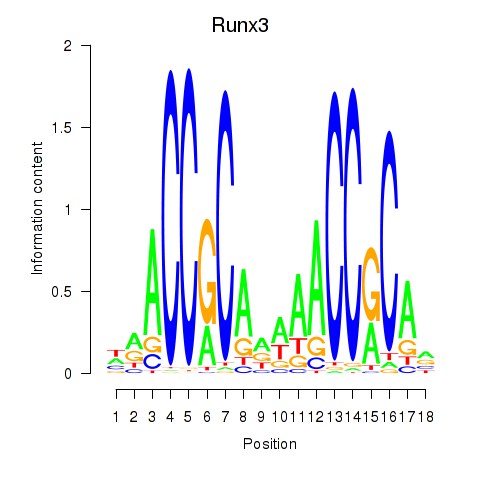
Results for Runx3
Z-value: 1.37
Transcription factors associated with Runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx3
|
ENSMUSG00000070691.11 | Runx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx3 | mm39_v1_chr4_+_134847949_134847973 | 0.76 | 9.3e-08 | Click! |
Activity profile of Runx3 motif
Sorted Z-values of Runx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_20368029 | 5.76 |
ENSMUST00000235280.2
|
Anxa1
|
annexin A1 |
| chr7_+_126690525 | 5.61 |
ENSMUST00000056288.7
ENSMUST00000206102.2 |
AI467606
|
expressed sequence AI467606 |
| chr13_-_22225527 | 5.57 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr15_-_78739717 | 5.51 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr17_+_27775637 | 4.23 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_27775613 | 4.19 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_27775471 | 4.10 |
ENSMUST00000118599.9
ENSMUST00000232265.2 ENSMUST00000232013.2 ENSMUST00000114888.11 ENSMUST00000231874.2 ENSMUST00000119486.9 ENSMUST00000231825.2 ENSMUST00000231866.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr13_-_59917569 | 4.05 |
ENSMUST00000057115.7
|
Isca1
|
iron-sulfur cluster assembly 1 |
| chr4_-_120427449 | 3.82 |
ENSMUST00000030381.8
|
Ctps
|
cytidine 5'-triphosphate synthase |
| chr2_+_4022537 | 3.56 |
ENSMUST00000177457.8
|
Frmd4a
|
FERM domain containing 4A |
| chr12_+_26519203 | 3.43 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr17_+_28547445 | 3.17 |
ENSMUST00000042334.16
|
Rpl10a
|
ribosomal protein L10A |
| chr17_+_28547533 | 2.96 |
ENSMUST00000233427.2
ENSMUST00000233937.2 |
Rpl10a
|
ribosomal protein L10A |
| chr4_-_116484675 | 2.94 |
ENSMUST00000081182.5
ENSMUST00000030457.12 |
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr9_+_106306736 | 2.90 |
ENSMUST00000098994.7
ENSMUST00000059802.7 ENSMUST00000213448.2 ENSMUST00000217081.2 |
Rpl29
|
ribosomal protein L29 |
| chr17_+_28547548 | 2.90 |
ENSMUST00000233895.2
ENSMUST00000232867.2 |
Rpl10a
|
ribosomal protein L10A |
| chr10_+_76367427 | 2.85 |
ENSMUST00000048678.7
|
Lss
|
lanosterol synthase |
| chr9_-_107971640 | 2.76 |
ENSMUST00000081309.13
ENSMUST00000191985.2 |
Apeh
|
acylpeptide hydrolase |
| chr11_-_5828251 | 2.46 |
ENSMUST00000102922.10
|
Pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr13_-_23735822 | 2.44 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr13_-_22016364 | 2.40 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr9_+_69919822 | 2.20 |
ENSMUST00000118198.8
ENSMUST00000119905.8 ENSMUST00000119413.8 ENSMUST00000140305.8 ENSMUST00000122087.8 |
Gtf2a2
|
general transcription factor II A, 2 |
| chr10_-_62438040 | 2.14 |
ENSMUST00000045866.9
|
Ddx21
|
DExD box helicase 21 |
| chr9_-_107971729 | 2.11 |
ENSMUST00000193254.6
|
Apeh
|
acylpeptide hydrolase |
| chr9_+_106306598 | 2.00 |
ENSMUST00000150576.8
|
Rpl29
|
ribosomal protein L29 |
| chr13_+_51799268 | 1.87 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr5_+_33176160 | 1.64 |
ENSMUST00000019109.8
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr13_-_51888737 | 1.53 |
ENSMUST00000110039.2
|
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr18_-_34784746 | 1.52 |
ENSMUST00000025228.12
ENSMUST00000133181.2 |
Cdc23
|
CDC23 cell division cycle 23 |
| chr17_+_36190662 | 1.48 |
ENSMUST00000025292.15
|
Dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr11_-_70300836 | 1.43 |
ENSMUST00000019065.10
ENSMUST00000135148.2 |
Pelp1
|
proline, glutamic acid and leucine rich protein 1 |
| chr11_-_100098333 | 1.40 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr11_-_70111796 | 1.29 |
ENSMUST00000060010.3
ENSMUST00000190533.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr7_+_28508220 | 1.28 |
ENSMUST00000172529.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr13_-_21934675 | 1.20 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr7_-_44624165 | 1.19 |
ENSMUST00000212836.2
ENSMUST00000212255.2 ENSMUST00000063761.8 |
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr5_+_115149170 | 1.17 |
ENSMUST00000031530.9
|
Sppl3
|
signal peptide peptidase 3 |
| chr7_-_25516041 | 1.08 |
ENSMUST00000043314.10
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr11_-_84807164 | 1.08 |
ENSMUST00000103195.5
|
Znhit3
|
zinc finger, HIT type 3 |
| chr2_-_151583155 | 0.95 |
ENSMUST00000042452.11
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr4_-_117749191 | 0.87 |
ENSMUST00000030265.4
|
Dph2
|
DPH2 homolog |
| chr5_+_121535999 | 0.84 |
ENSMUST00000042163.15
|
Naa25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
| chr9_+_121548237 | 0.81 |
ENSMUST00000035112.13
ENSMUST00000182311.8 |
Nktr
|
natural killer tumor recognition sequence |
| chr10_+_42378193 | 0.80 |
ENSMUST00000105499.2
|
Snx3
|
sorting nexin 3 |
| chr7_+_78432867 | 0.79 |
ENSMUST00000032840.5
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr4_-_70328659 | 0.76 |
ENSMUST00000144099.8
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chr6_-_100648086 | 0.75 |
ENSMUST00000089245.7
ENSMUST00000113312.9 ENSMUST00000170667.8 |
Shq1
|
SHQ1 homolog (S. cerevisiae) |
| chr13_-_23882437 | 0.71 |
ENSMUST00000102967.3
|
H4c3
|
H4 clustered histone 3 |
| chr1_+_74317709 | 0.68 |
ENSMUST00000077985.4
|
Gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr6_-_136781406 | 0.67 |
ENSMUST00000179285.3
|
H4f16
|
H4 histone 16 |
| chrX_-_72502595 | 0.66 |
ENSMUST00000033737.15
ENSMUST00000077243.5 |
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr10_+_79590910 | 0.65 |
ENSMUST00000219981.2
ENSMUST00000219228.2 ENSMUST00000020577.4 |
Fgf22
|
fibroblast growth factor 22 |
| chr7_+_140796537 | 0.63 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr6_+_124690060 | 0.58 |
ENSMUST00000130279.2
|
Phb2
|
prohibitin 2 |
| chr16_-_20972750 | 0.51 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr11_-_70111602 | 0.44 |
ENSMUST00000141290.2
ENSMUST00000159867.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr3_+_95566082 | 0.43 |
ENSMUST00000037947.15
ENSMUST00000178686.2 |
Mcl1
|
myeloid cell leukemia sequence 1 |
| chr1_+_82891043 | 0.42 |
ENSMUST00000220768.2
|
A030005L19Rik
|
RIKEN cDNA A030005L19 gene |
| chr17_-_45744637 | 0.40 |
ENSMUST00000024727.10
|
Cdc5l
|
cell division cycle 5-like (S. pombe) |
| chrX_-_97934387 | 0.39 |
ENSMUST00000113826.8
ENSMUST00000033560.9 ENSMUST00000142267.2 |
Ophn1
|
oligophrenin 1 |
| chr11_-_32217547 | 0.37 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr4_-_133225849 | 0.35 |
ENSMUST00000125541.2
|
Trnp1
|
TMF1-regulated nuclear protein 1 |
| chr10_-_116417333 | 0.34 |
ENSMUST00000218744.2
ENSMUST00000105267.8 ENSMUST00000105265.8 ENSMUST00000167706.8 ENSMUST00000168036.8 ENSMUST00000169921.8 ENSMUST00000020374.6 |
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr7_-_141674466 | 0.32 |
ENSMUST00000209890.2
|
Gm45618
|
predicted gene 45618 |
| chr9_+_110361561 | 0.32 |
ENSMUST00000153838.8
|
Setd2
|
SET domain containing 2 |
| chr18_-_61919707 | 0.28 |
ENSMUST00000120472.2
|
Afap1l1
|
actin filament associated protein 1-like 1 |
| chr10_+_127919142 | 0.27 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr2_-_53081199 | 0.26 |
ENSMUST00000239398.2
ENSMUST00000076313.14 ENSMUST00000210789.3 |
Prpf40a
|
pre-mRNA processing factor 40A |
| chr2_-_50186690 | 0.25 |
ENSMUST00000144143.8
ENSMUST00000102769.11 ENSMUST00000133768.2 |
Mmadhc
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr13_-_115238427 | 0.23 |
ENSMUST00000224997.2
ENSMUST00000061673.9 |
Gm49395
Itga1
|
predicted gene, 49395 integrin alpha 1 |
| chr9_+_121548469 | 0.21 |
ENSMUST00000182225.8
|
Nktr
|
natural killer tumor recognition sequence |
| chr13_+_21919225 | 0.17 |
ENSMUST00000087714.6
|
H4c11
|
H4 clustered histone 11 |
| chr2_+_37080286 | 0.16 |
ENSMUST00000218602.2
|
Olfr365
|
olfactory receptor 365 |
| chr9_-_78389006 | 0.16 |
ENSMUST00000042235.15
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr18_+_37939442 | 0.14 |
ENSMUST00000076807.7
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr18_+_47245204 | 0.14 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr15_+_40518414 | 0.14 |
ENSMUST00000053467.6
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr12_-_98225676 | 0.04 |
ENSMUST00000021390.9
|
Galc
|
galactosylceramidase |
| chr3_-_7678796 | 0.03 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr14_-_98406977 | 0.03 |
ENSMUST00000071533.13
ENSMUST00000069334.8 |
Dach1
|
dachshund family transcription factor 1 |
| chr7_+_140796559 | 0.01 |
ENSMUST00000148975.3
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr9_+_58395850 | 0.01 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr1_+_40505136 | 0.00 |
ENSMUST00000087983.8
ENSMUST00000195684.6 ENSMUST00000108044.4 |
Il18r1
|
interleukin 18 receptor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 1.1 | 3.4 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.5 | 1.6 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.5 | 4.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.4 | 1.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.3 | 2.9 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 0.8 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 9.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.3 | 3.8 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.3 | 0.8 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 12.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.2 | 2.5 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.2 | 1.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.9 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 1.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.3 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.6 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 3.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 4.9 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 1.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 4.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 2.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 1.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 1.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.4 | GO:0072401 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 1.3 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 12.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 5.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 2.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 2.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 4.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 9.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.2 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.8 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.8 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.4 | GO:0005662 | Prp19 complex(GO:0000974) DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 3.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 4.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.1 | 3.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 1.0 | 3.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.6 | 5.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.6 | 5.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 1.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 4.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 1.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 2.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.7 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 1.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 4.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 2.8 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 14.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 2.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 2.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 12.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 2.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 14.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.5 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.1 | 1.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 5.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.0 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |