Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Rxrb
Z-value: 1.28
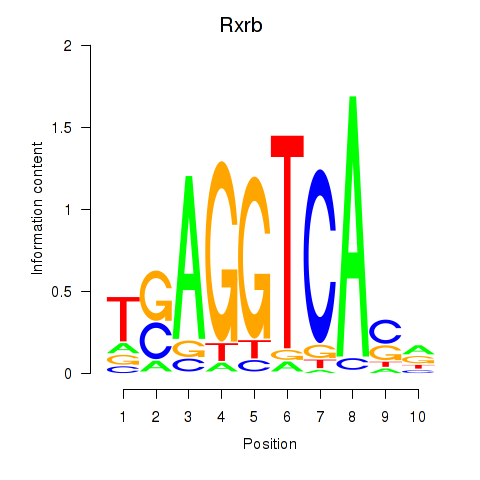
Transcription factors associated with Rxrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxrb
|
ENSMUSG00000039656.18 | Rxrb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrb | mm39_v1_chr17_+_34251041_34251170 | -0.81 | 3.1e-09 | Click! |
Activity profile of Rxrb motif
Sorted Z-values of Rxrb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxrb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_180021218 | 4.04 |
ENSMUST00000159914.8
|
Coq8a
|
coenzyme Q8A |
| chr1_+_133291302 | 3.80 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr17_+_25097199 | 3.18 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr1_-_140111138 | 2.85 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 2.71 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr1_+_167445815 | 2.60 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr15_-_76193955 | 2.56 |
ENSMUST00000210024.2
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr7_-_105249308 | 2.45 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr3_-_89300936 | 2.28 |
ENSMUST00000124783.8
ENSMUST00000126027.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr6_-_116050081 | 2.06 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr2_+_71884943 | 1.98 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_+_75084609 | 1.94 |
ENSMUST00000102514.4
|
Rtn4rl1
|
reticulon 4 receptor-like 1 |
| chr6_+_129510331 | 1.92 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr5_-_116560916 | 1.88 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr4_+_41760454 | 1.78 |
ENSMUST00000108040.8
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr9_-_106769069 | 1.61 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr6_+_129510145 | 1.60 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr6_+_129510117 | 1.53 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr11_-_53321606 | 1.50 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr7_-_141014336 | 1.50 |
ENSMUST00000136354.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr9_-_106769131 | 1.46 |
ENSMUST00000159283.8
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr7_-_141014445 | 1.43 |
ENSMUST00000133021.2
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr10_-_78300802 | 1.41 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr7_-_126522014 | 1.33 |
ENSMUST00000134134.3
ENSMUST00000119781.8 ENSMUST00000121612.4 |
Tmem219
|
transmembrane protein 219 |
| chr14_+_6226418 | 1.31 |
ENSMUST00000112625.9
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr7_-_141014192 | 1.31 |
ENSMUST00000201127.5
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr1_+_133109059 | 1.26 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr10_-_67972401 | 1.18 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr8_-_68363564 | 1.16 |
ENSMUST00000093468.12
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr16_-_23339548 | 1.16 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr10_+_61556371 | 1.16 |
ENSMUST00000080099.6
|
Aifm2
|
apoptosis-inducing factor, mitochondrion-associated 2 |
| chr7_-_30755007 | 1.15 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr6_-_124942366 | 1.13 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chrX_+_72760183 | 1.09 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chrX_-_99456185 | 1.06 |
ENSMUST00000033567.15
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr2_-_6217844 | 1.04 |
ENSMUST00000042658.5
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr11_+_97697128 | 1.02 |
ENSMUST00000138919.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr8_+_77626400 | 1.02 |
ENSMUST00000109913.9
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr11_-_120348762 | 1.01 |
ENSMUST00000137632.2
ENSMUST00000044007.3 |
Oxld1
|
oxidoreductase like domain containing 1 |
| chr6_-_124840824 | 1.01 |
ENSMUST00000046893.10
ENSMUST00000204667.2 |
Gpr162
|
G protein-coupled receptor 162 |
| chr16_-_30207348 | 0.98 |
ENSMUST00000061350.13
ENSMUST00000100013.9 |
Atp13a3
|
ATPase type 13A3 |
| chr4_+_152262583 | 0.96 |
ENSMUST00000075363.10
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr6_+_22288220 | 0.95 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr3_+_32791139 | 0.94 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr1_-_190897012 | 0.92 |
ENSMUST00000171798.2
|
Fam71a
|
family with sequence similarity 71, member A |
| chr13_+_113931155 | 0.92 |
ENSMUST00000224068.2
|
Arl15
|
ADP-ribosylation factor-like 15 |
| chr6_-_124942170 | 0.90 |
ENSMUST00000148485.2
ENSMUST00000129976.8 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr11_-_95037089 | 0.90 |
ENSMUST00000021241.8
|
Dlx4
|
distal-less homeobox 4 |
| chr11_+_97697328 | 0.89 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1 |
| chr19_-_6957789 | 0.87 |
ENSMUST00000070878.9
ENSMUST00000177752.9 |
Fkbp2
|
FK506 binding protein 2 |
| chr16_+_31482745 | 0.87 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr1_+_84817547 | 0.86 |
ENSMUST00000097672.4
|
Fbxo36
|
F-box protein 36 |
| chr5_+_73071897 | 0.85 |
ENSMUST00000200785.2
|
Slain2
|
SLAIN motif family, member 2 |
| chr9_+_55116209 | 0.84 |
ENSMUST00000034859.15
|
Fbxo22
|
F-box protein 22 |
| chr14_-_31552335 | 0.81 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chrX_+_72760318 | 0.80 |
ENSMUST00000114461.3
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr6_-_124942457 | 0.78 |
ENSMUST00000112439.9
|
Cops7a
|
COP9 signalosome subunit 7A |
| chr11_+_51541728 | 0.77 |
ENSMUST00000117859.8
ENSMUST00000064493.6 |
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr11_+_72686990 | 0.75 |
ENSMUST00000069395.7
ENSMUST00000172220.8 |
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr19_+_11747721 | 0.74 |
ENSMUST00000167199.3
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr1_-_52539395 | 0.73 |
ENSMUST00000186764.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr17_-_28039419 | 0.73 |
ENSMUST00000233508.2
ENSMUST00000075076.6 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr1_-_156936197 | 0.72 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr11_+_97692738 | 0.72 |
ENSMUST00000127033.9
|
Lasp1
|
LIM and SH3 protein 1 |
| chr10_-_8761777 | 0.71 |
ENSMUST00000015449.6
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr11_+_100960838 | 0.71 |
ENSMUST00000001802.10
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chr19_+_11863929 | 0.70 |
ENSMUST00000217281.2
|
Olfr1420
|
olfactory receptor 1420 |
| chr15_-_101471324 | 0.69 |
ENSMUST00000023714.5
|
Krt90
|
keratin 90 |
| chr3_+_129326004 | 0.67 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr2_-_48839276 | 0.67 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr15_-_73702827 | 0.67 |
ENSMUST00000238402.2
|
Mroh5
|
maestro heat-like repeat family member 5 |
| chr17_-_28039506 | 0.65 |
ENSMUST00000114859.9
ENSMUST00000233533.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr3_+_129326285 | 0.64 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr4_+_155916087 | 0.64 |
ENSMUST00000105592.8
ENSMUST00000105591.2 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr19_+_45064539 | 0.63 |
ENSMUST00000026234.5
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr2_-_48839218 | 0.63 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr5_+_73071607 | 0.62 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr12_+_33364288 | 0.61 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr15_-_82238432 | 0.60 |
ENSMUST00000230494.2
ENSMUST00000023085.7 |
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr5_+_73071695 | 0.59 |
ENSMUST00000143829.5
|
Slain2
|
SLAIN motif family, member 2 |
| chr6_-_88423464 | 0.59 |
ENSMUST00000204459.3
ENSMUST00000203213.3 ENSMUST00000205179.3 ENSMUST00000165242.4 |
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr11_-_101875575 | 0.59 |
ENSMUST00000176261.2
ENSMUST00000143177.2 ENSMUST00000003612.13 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr1_+_6557455 | 0.59 |
ENSMUST00000140079.8
ENSMUST00000131494.8 |
St18
|
suppression of tumorigenicity 18 |
| chr2_-_122144125 | 0.58 |
ENSMUST00000147788.8
ENSMUST00000154412.2 ENSMUST00000110537.8 ENSMUST00000148417.8 |
Duoxa1
|
dual oxidase maturation factor 1 |
| chr14_-_21791544 | 0.58 |
ENSMUST00000184703.8
ENSMUST00000183698.8 ENSMUST00000119866.9 |
Dusp13
|
dual specificity phosphatase 13 |
| chr1_-_13197387 | 0.58 |
ENSMUST00000047577.7
|
Prdm14
|
PR domain containing 14 |
| chrX_+_85235370 | 0.57 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr16_+_23338960 | 0.56 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr7_-_119694400 | 0.55 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr3_+_122298308 | 0.55 |
ENSMUST00000199358.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr15_+_38219447 | 0.54 |
ENSMUST00000081966.5
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr2_-_76478336 | 0.54 |
ENSMUST00000002808.7
|
Prkra
|
protein kinase, interferon inducible double stranded RNA dependent activator |
| chr17_-_67939702 | 0.54 |
ENSMUST00000097290.4
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr2_+_48839505 | 0.54 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr15_+_76152276 | 0.53 |
ENSMUST00000074173.4
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr11_-_97520511 | 0.53 |
ENSMUST00000052281.6
|
Epop
|
elongin BC and polycomb repressive complex 2 associated protein |
| chr16_-_44153288 | 0.53 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr2_-_25104628 | 0.52 |
ENSMUST00000043774.11
ENSMUST00000114363.2 |
Stpg3
|
sperm tail PG rich repeat containing 3 |
| chr12_+_119909692 | 0.51 |
ENSMUST00000058644.15
|
Tmem196
|
transmembrane protein 196 |
| chr10_-_128657445 | 0.51 |
ENSMUST00000217685.2
ENSMUST00000026409.5 ENSMUST00000219215.2 ENSMUST00000219524.2 |
Ormdl2
|
ORM1-like 2 (S. cerevisiae) |
| chr2_-_150934270 | 0.50 |
ENSMUST00000109890.2
|
Gm14151
|
predicted gene 14151 |
| chr5_+_3853160 | 0.50 |
ENSMUST00000080085.9
ENSMUST00000200386.5 |
Krit1
|
KRIT1, ankyrin repeat containing |
| chr9_+_45313913 | 0.50 |
ENSMUST00000214257.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr2_+_174602574 | 0.50 |
ENSMUST00000140908.2
|
Edn3
|
endothelin 3 |
| chr8_-_13612397 | 0.49 |
ENSMUST00000187391.7
ENSMUST00000134023.9 ENSMUST00000151400.10 |
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene |
| chr17_-_18075450 | 0.48 |
ENSMUST00000003762.8
|
Has1
|
hyaluronan synthase 1 |
| chr17_+_28794615 | 0.47 |
ENSMUST00000232862.2
ENSMUST00000080780.8 |
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr7_+_45164042 | 0.47 |
ENSMUST00000210868.2
|
Tulp2
|
tubby-like protein 2 |
| chr8_+_110220614 | 0.46 |
ENSMUST00000034162.8
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr17_-_36014863 | 0.46 |
ENSMUST00000148065.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr12_-_98703664 | 0.45 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr17_+_28794727 | 0.45 |
ENSMUST00000232974.2
|
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr7_+_45164005 | 0.45 |
ENSMUST00000210532.2
ENSMUST00000085331.14 ENSMUST00000210299.2 |
Tulp2
|
tubby-like protein 2 |
| chr16_-_44153498 | 0.44 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr11_-_3672188 | 0.44 |
ENSMUST00000102950.10
ENSMUST00000101632.4 |
Osbp2
|
oxysterol binding protein 2 |
| chr11_+_101875095 | 0.44 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr5_-_25200745 | 0.44 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr10_-_17823736 | 0.43 |
ENSMUST00000037879.8
|
Heca
|
hdc homolog, cell cycle regulator |
| chr17_-_28039588 | 0.42 |
ENSMUST00000114863.10
ENSMUST00000233131.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr11_-_94376158 | 0.42 |
ENSMUST00000041705.8
|
Spata20
|
spermatogenesis associated 20 |
| chr4_-_20778847 | 0.41 |
ENSMUST00000102998.4
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr4_-_20778530 | 0.41 |
ENSMUST00000119374.8
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr1_+_75456173 | 0.41 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr8_+_12623016 | 0.41 |
ENSMUST00000210276.2
ENSMUST00000010579.8 ENSMUST00000209428.2 |
Spaca7
|
sperm acrosome associated 7 |
| chr11_+_40624466 | 0.41 |
ENSMUST00000020578.11
|
Nudcd2
|
NudC domain containing 2 |
| chr2_+_121279842 | 0.40 |
ENSMUST00000110615.8
ENSMUST00000099475.12 |
Serf2
|
small EDRK-rich factor 2 |
| chr9_-_50571080 | 0.40 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr2_+_150940165 | 0.38 |
ENSMUST00000109888.2
|
Gm14147
|
predicted gene 14147 |
| chr17_-_24863956 | 0.37 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chrX_-_100307592 | 0.37 |
ENSMUST00000101358.3
|
Gm614
|
predicted gene 614 |
| chr12_-_111679379 | 0.36 |
ENSMUST00000160825.2
ENSMUST00000162953.2 |
Bag5
|
BCL2-associated athanogene 5 |
| chr11_+_72687080 | 0.36 |
ENSMUST00000207107.2
|
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr7_-_4774277 | 0.36 |
ENSMUST00000174409.2
|
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr11_-_104332528 | 0.36 |
ENSMUST00000106971.2
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr4_-_58912678 | 0.36 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr16_-_23339329 | 0.35 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr5_-_74863514 | 0.34 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr14_+_53061814 | 0.34 |
ENSMUST00000103648.4
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr14_+_58308004 | 0.34 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr19_-_45772230 | 0.34 |
ENSMUST00000235448.2
|
Oga
|
O-GlcNAcase |
| chr17_-_24863907 | 0.33 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr3_-_94693740 | 0.33 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
| chr6_+_90279064 | 0.33 |
ENSMUST00000167550.4
|
BC048671
|
cDNA sequence BC048671 |
| chr15_+_92059224 | 0.32 |
ENSMUST00000068378.6
|
Cntn1
|
contactin 1 |
| chr1_+_133254950 | 0.32 |
ENSMUST00000178033.5
|
Kiss1
|
KiSS-1 metastasis-suppressor |
| chr11_-_78056347 | 0.32 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr17_-_37178079 | 0.32 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chrX_-_7774074 | 0.32 |
ENSMUST00000085330.5
|
Gm10491
|
predicted gene 10491 |
| chr13_+_73911797 | 0.31 |
ENSMUST00000017900.9
|
Slc12a7
|
solute carrier family 12, member 7 |
| chr4_+_57821050 | 0.31 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr7_+_16693604 | 0.31 |
ENSMUST00000038163.8
|
Pnmal1
|
PNMA-like 1 |
| chr6_+_71520855 | 0.31 |
ENSMUST00000204535.2
ENSMUST00000065364.5 ENSMUST00000204199.2 |
Chmp3
|
charged multivesicular body protein 3 |
| chr7_+_105289655 | 0.30 |
ENSMUST00000058333.10
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr17_+_21503204 | 0.30 |
ENSMUST00000236545.2
ENSMUST00000235527.2 |
Vmn1r236
|
vomeronasal 1 receptor 236 |
| chr6_+_71520781 | 0.29 |
ENSMUST00000059462.12
|
Chmp3
|
charged multivesicular body protein 3 |
| chr4_-_139695337 | 0.28 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr1_-_162305573 | 0.28 |
ENSMUST00000086074.12
ENSMUST00000070330.14 |
Dnm3
|
dynamin 3 |
| chr15_-_5273645 | 0.27 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr17_-_66756710 | 0.27 |
ENSMUST00000086693.12
ENSMUST00000097291.10 |
Mtcl1
|
microtubule crosslinking factor 1 |
| chr2_-_151318073 | 0.27 |
ENSMUST00000080132.3
|
4921509C19Rik
|
RIKEN cDNA 4921509C19 gene |
| chr15_-_5273659 | 0.26 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr11_+_40624763 | 0.26 |
ENSMUST00000127382.2
|
Nudcd2
|
NudC domain containing 2 |
| chr1_+_85949827 | 0.26 |
ENSMUST00000131151.3
|
Spata3
|
spermatogenesis associated 3 |
| chr4_+_148085179 | 0.26 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr7_+_137039309 | 0.26 |
ENSMUST00000064404.8
ENSMUST00000211496.2 ENSMUST00000209696.2 |
Glrx3
|
glutaredoxin 3 |
| chr6_+_119213468 | 0.24 |
ENSMUST00000238905.2
ENSMUST00000037434.13 |
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr1_+_131616504 | 0.24 |
ENSMUST00000129905.2
|
Rab7b
|
RAB7B, member RAS oncogene family |
| chr7_+_101879176 | 0.24 |
ENSMUST00000120119.9
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr1_+_131678223 | 0.23 |
ENSMUST00000147800.2
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr15_+_98953776 | 0.23 |
ENSMUST00000229268.2
|
Prph
|
peripherin |
| chr16_+_24540599 | 0.22 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr12_+_111679689 | 0.22 |
ENSMUST00000040519.12
ENSMUST00000163220.10 ENSMUST00000162316.2 |
Coa8
|
cytochrome c oxidase assembly factor 8 |
| chr7_-_104835219 | 0.21 |
ENSMUST00000214712.2
ENSMUST00000217432.2 ENSMUST00000209409.3 |
Olfr685
|
olfactory receptor 685 |
| chr3_+_107009896 | 0.20 |
ENSMUST00000196403.2
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr2_-_153712996 | 0.20 |
ENSMUST00000028982.5
|
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr16_-_10608766 | 0.20 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr3_+_89970088 | 0.20 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr2_+_174602412 | 0.20 |
ENSMUST00000029030.9
|
Edn3
|
endothelin 3 |
| chr1_-_38168801 | 0.19 |
ENSMUST00000195383.2
|
Rev1
|
REV1, DNA directed polymerase |
| chr19_-_6036171 | 0.19 |
ENSMUST00000041827.8
|
Slc22a20
|
solute carrier family 22 (organic anion transporter), member 20 |
| chr3_-_97731826 | 0.19 |
ENSMUST00000200232.3
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr2_-_136229849 | 0.19 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr14_+_54713703 | 0.19 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr11_-_99511257 | 0.18 |
ENSMUST00000073853.3
|
Gm11562
|
predicted gene 11562 |
| chr1_-_171359228 | 0.18 |
ENSMUST00000168184.2
|
Itln1
|
intelectin 1 (galactofuranose binding) |
| chr4_-_106585127 | 0.18 |
ENSMUST00000106770.8
ENSMUST00000145044.2 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chr12_+_86781141 | 0.18 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr14_+_47710574 | 0.17 |
ENSMUST00000228740.2
|
Fbxo34
|
F-box protein 34 |
| chr9_-_77159152 | 0.17 |
ENSMUST00000183955.8
|
Mlip
|
muscular LMNA-interacting protein |
| chr19_+_42040681 | 0.16 |
ENSMUST00000164518.4
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr5_-_66672158 | 0.16 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr12_+_86781154 | 0.16 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr7_+_15909010 | 0.16 |
ENSMUST00000176342.8
ENSMUST00000177540.8 |
Meis3
|
Meis homeobox 3 |
| chr9_-_77159111 | 0.16 |
ENSMUST00000184322.8
ENSMUST00000184316.2 |
Mlip
|
muscular LMNA-interacting protein |
| chr5_-_137623331 | 0.16 |
ENSMUST00000031732.14
|
Fbxo24
|
F-box protein 24 |
| chr7_-_133966588 | 0.15 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr12_+_91367764 | 0.15 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr10_+_93476903 | 0.15 |
ENSMUST00000020204.5
|
Ntn4
|
netrin 4 |
| chr4_+_57845240 | 0.13 |
ENSMUST00000102903.8
ENSMUST00000107598.9 |
Pakap
|
paralemmin A kinase anchor protein |
| chr8_+_72860884 | 0.13 |
ENSMUST00000210435.4
|
Olfr374
|
olfactory receptor 374 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.6 | 5.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.6 | 2.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.5 | 1.9 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.5 | 1.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.4 | 2.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 3.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 2.5 | GO:0015886 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 2.3 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 5.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 0.5 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 1.0 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.2 | 0.9 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.2 | 1.5 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.2 | 0.6 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 1.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 0.5 | GO:2000388 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.2 | 1.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 0.5 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 2.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.6 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 4.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.5 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 2.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.7 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.1 | 1.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.2 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 1.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 1.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.3 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 1.8 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.6 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.6 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.9 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 2.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.8 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 2.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.6 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 1.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 2.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:1903044 | protein transport into membrane raft(GO:0032596) protein localization to plasma membrane raft(GO:0044860) protein transport into plasma membrane raft(GO:0044861) protein localization to membrane raft(GO:1903044) |
| 0.0 | 0.1 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.0 | 0.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0072257 | condensed mesenchymal cell proliferation(GO:0072137) metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.5 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 3.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 5.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 0.5 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 2.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 1.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 6.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.8 | 5.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.5 | 1.4 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.4 | 1.8 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.4 | 2.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 0.4 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.3 | 1.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 2.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 4.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.3 | 1.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.3 | 1.3 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 2.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 5.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 1.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 1.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 1.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 3.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 4.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.7 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.6 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 3.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 4.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.0 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |